Web Sites & Databases
We produce a wide variety of websites for use by researchers at the institute, around the UK and the world at large. You’ll find brief descriptions of many of them on this page, along with a links to the actual sites. Shortcut links to take you directly to a site can be found in the navigation menu.
UK Barley Genome Sequencing
 The UK Barley Genome Sequencing work is part of a wider effort to help gain a better understanding of this important crop with the aim of producing varieties that are more resilient, maintain yield and resist pests and disease. Barley is the fourth ranked cereal crop produced in the world and the second in terms of harvest in UK agriculture. It is an essential component of the whisky and beer industry that is worth more than £20 billion alone to the UK economy. In addition, it is a major contributor to animal feed and is still a significant calorie and roughage source in the human diet in several parts of the world.
The UK Barley Genome Sequencing work is part of a wider effort to help gain a better understanding of this important crop with the aim of producing varieties that are more resilient, maintain yield and resist pests and disease. Barley is the fourth ranked cereal crop produced in the world and the second in terms of harvest in UK agriculture. It is an essential component of the whisky and beer industry that is worth more than £20 billion alone to the UK economy. In addition, it is a major contributor to animal feed and is still a significant calorie and roughage source in the human diet in several parts of the world.
|
More information: |
http://barleygenome.org.uk |
|
Primary developers: |
Paulo Flores |
Germinate
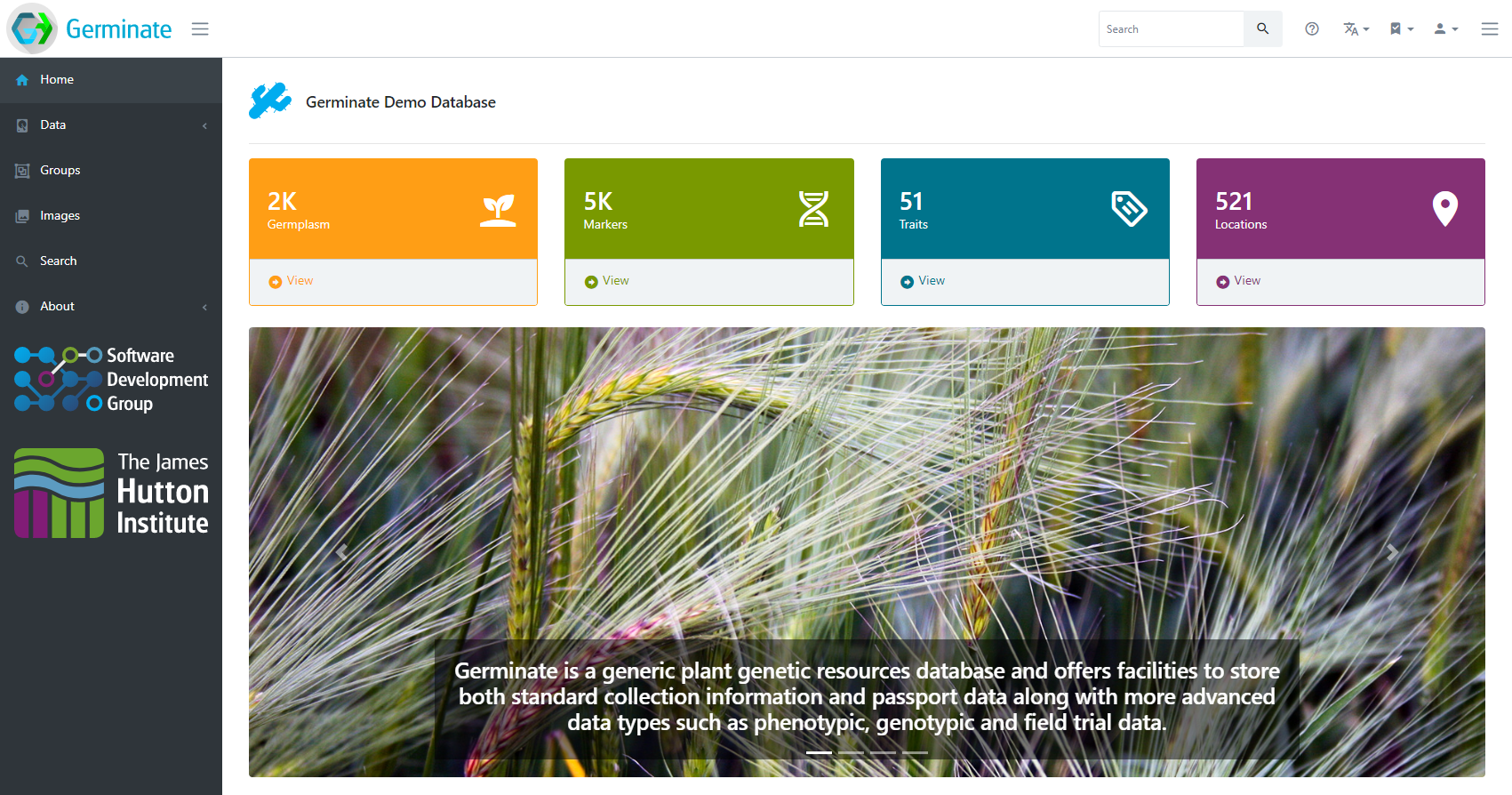 The latest version of Germinate adds even more new features, visualizations and customization options. We now offer Docker containers for easier setup and better documentation to get started. The underlying database is fully compatible with Germinate 3 but has had some tweaks made in order to increase the functionality that we can offer. The first release of the new Germinate was in support of the Crop Wild Relatives project (www.cwrdiversity.org) in collaboration with the Crop Trust (www.croptrust.org) in Germany.
The latest version of Germinate adds even more new features, visualizations and customization options. We now offer Docker containers for easier setup and better documentation to get started. The underlying database is fully compatible with Germinate 3 but has had some tweaks made in order to increase the functionality that we can offer. The first release of the new Germinate was in support of the Crop Wild Relatives project (www.cwrdiversity.org) in collaboration with the Crop Trust (www.croptrust.org) in Germany.
|
More information: |
https://ics.hutton.ac.uk/get-germinate |
|
Primary developers: |
Sebastian Raubach and Paul Shaw |
Germinate 3
 Germinate 3 builds on the Germinate 2 platform but allows us to include many new additional features. Germinate 3 uses a MySQL, Java and Google Web Toolkit (GWT) based solution. The underlying database is fully compatible with Germinate 2 but has had some tweaks made in order to increase the functionality that we can offer. GWT allows us to offer a rich web-based interface to our datasets and is a considerable upgrade to its predecessor. The first release of Germinate 3 was in support of the Seeds of Discovery project (www.seedsofdiscovery.org) in collaboration with CIMMYT (www.cimmyt.org) in Mexico.
Germinate 3 builds on the Germinate 2 platform but allows us to include many new additional features. Germinate 3 uses a MySQL, Java and Google Web Toolkit (GWT) based solution. The underlying database is fully compatible with Germinate 2 but has had some tweaks made in order to increase the functionality that we can offer. GWT allows us to offer a rich web-based interface to our datasets and is a considerable upgrade to its predecessor. The first release of Germinate 3 was in support of the Seeds of Discovery project (www.seedsofdiscovery.org) in collaboration with CIMMYT (www.cimmyt.org) in Mexico.
|
More information: |
https://ics.hutton.ac.uk/get-germinate |
|
Primary developers: |
Paul Shaw and Sebastian Raubach |
Germinate 2
 Germinate 2 is a generic plant data management system implemented in MySQL and designed to hold passport data and a range of additional data types including molecular markers and phenotypic data.
Germinate 2 is a generic plant data management system implemented in MySQL and designed to hold passport data and a range of additional data types including molecular markers and phenotypic data.
Germinate 2 used a Perl based web-interface to allow users to browse, query and export data.
Its use of a top level Accessions entry ID allows association and querying between disparate sets of data associated with an accession.
The database is implemented in the public domain with MySQL relational database management system and it is freely available under the terms of the GNU public license. We envisage that this will form an ideal platform to enable us and other groups to develop a range of standard interfaces and analytical tools to interact with the underlying database.
While there are a number of resources that use Germinate 2 it is no longer under active development and we have moved to the Germinate 3 platform which takes many of the features that made Germinate 2 so popular and has added additional functionality.
|
More information: |
https://ics.hutton.ac.uk/get-germinate |
|
Primary developers: |
Paul Shaw |
Barley SNP Database
 This online database contains information from a project at the James Hutton Institute to mine wheat and barley genes associated with abiotic stress for SNPs which were then mapped in barley crosses. The reference sequence for each gene was an EST unigene from the HarvEST Barley Assembly 21, which had also been used to design Affymetrix probe sets for the Barley1 GeneChip. A selection of EST unigenes were analyzed for sequence polymorphisms in 8 diverse barley genotypes: OWB D, OWB R, Steptoe, Morex, Lina, HS92, Optic and Golden Promise. The database includes the aligned gene sequences in the 8 genotypes, the identified SNPs, the SNP mapping locations, and primers designed to those SNPs. Further details about the project can be seen here. An online abstract of the published paper can be seen here
This online database contains information from a project at the James Hutton Institute to mine wheat and barley genes associated with abiotic stress for SNPs which were then mapped in barley crosses. The reference sequence for each gene was an EST unigene from the HarvEST Barley Assembly 21, which had also been used to design Affymetrix probe sets for the Barley1 GeneChip. A selection of EST unigenes were analyzed for sequence polymorphisms in 8 diverse barley genotypes: OWB D, OWB R, Steptoe, Morex, Lina, HS92, Optic and Golden Promise. The database includes the aligned gene sequences in the 8 genotypes, the identified SNPs, the SNP mapping locations, and primers designed to those SNPs. Further details about the project can be seen here. An online abstract of the published paper can be seen here
|
More information: |
http://bioinf.hutton.ac.uk/barley_snpdb |
|
Primary developers: |
Linda Milne |
Barley Epigenome Browser
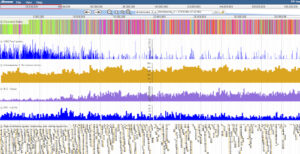 This resource is a collection of histone modification data for the whole barley seedling cv. Morex. Using chromatin immunoprecipitation followed by Next Generation Sequencing (ChIP-seq) nine histone modifications were surveyed in the barley genome. Chromatin states were also defined with ChromHMM and mapped to the genome. Gene and transposable element annotations are provided as tracks in the genome browser. This work was funded by BBSRC grant BB/I022899/1 “The diversity and evolution of the gene component of barley peri-centromeric heterochromatin” and was a collaboration with Prof Andy Flavell of the University of Dundee. Access the barley epigenome browser and associated data at the following URL.
This resource is a collection of histone modification data for the whole barley seedling cv. Morex. Using chromatin immunoprecipitation followed by Next Generation Sequencing (ChIP-seq) nine histone modifications were surveyed in the barley genome. Chromatin states were also defined with ChromHMM and mapped to the genome. Gene and transposable element annotations are provided as tracks in the genome browser. This work was funded by BBSRC grant BB/I022899/1 “The diversity and evolution of the gene component of barley peri-centromeric heterochromatin” and was a collaboration with Prof Andy Flavell of the University of Dundee. Access the barley epigenome browser and associated data at the following URL.
|
More information: |
https://ics.hutton.ac.uk/barley-epigenome/ |
|
Primary developers: |
Katie Baker and Iain Milne |
Plant snoRNA Database
 This resource brings together information from three independant computer-assisted searches of the Arabidopsis genome for box C/D snoRNA genes and from studies of ncRNAs.
This resource brings together information from three independant computer-assisted searches of the Arabidopsis genome for box C/D snoRNA genes and from studies of ncRNAs.
This resource was developed in collaboration with Professor John Brown (University of Dundee)
|
More information: |
http://bioinf.hutton.ac.uk/cgi-bin/plant_snorna/home |
|
Primary developers: |
Paul Shaw |
AtNoPDB
The Arabidopsis Nucleolar Protein database (AtNoPDB) provides information on 217 Arabidopsis proteins in comparison to human and yeast proteins.
|
More information: |
http://bioinf.hutton.ac.uk/cgi-bin/atnopdb/home |
|
Primary developers: |
Paul Shaw |
Wheat In-Situ Database
This resource was developed and implemented at SCRI to hold the data obtained from a joint BBSRC and Syngenta project for high-throughput wheat in situ hybridisation.
|
More information: |
http://bioinf.hutton.ac.uk/cgi-bin/insitu/home |
|
Primary developers: |
Paul Shaw |
OPTIRas
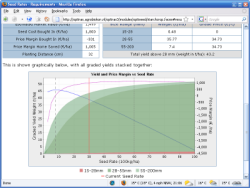 In collaboration with Dutch and German partners (under the umbrella of Agrobiokon), we developed several versions of a web-based decision support system for starch potato growers. The latest version – OPTIRas III – provides advanced decision support and e-learning/knowledge transfer for starch potato growers in the Netherlands and the Niedersachsen region of Northern Germany. OPTIRas provides five main areas of support covering cultivar selection, seed rates, PCN resistance, nutrients, and harvest damage, and the system is available in three languages (English, Dutch, and German). Note that this project is no longer active.
In collaboration with Dutch and German partners (under the umbrella of Agrobiokon), we developed several versions of a web-based decision support system for starch potato growers. The latest version – OPTIRas III – provides advanced decision support and e-learning/knowledge transfer for starch potato growers in the Netherlands and the Niedersachsen region of Northern Germany. OPTIRas provides five main areas of support covering cultivar selection, seed rates, PCN resistance, nutrients, and harvest damage, and the system is available in three languages (English, Dutch, and German). Note that this project is no longer active.
|
More information: |
https://ics.hutton.ac.uk/optiras |
|
Primary developers: |
Iain Milne and Micha Bayer |
