Barley epigenome
Barley epigenome browser
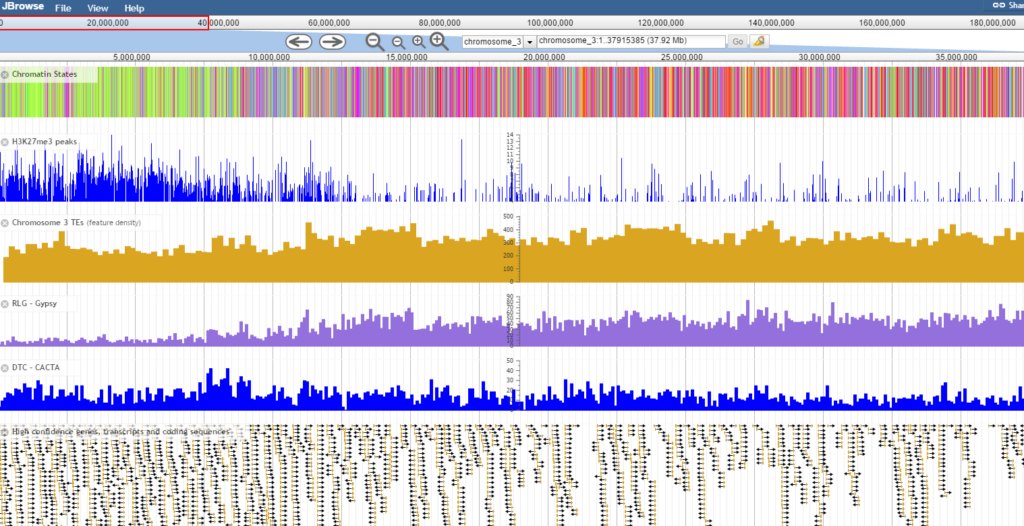
The barley epigenome browser is a JBrowse based genome browser for visualising barley histone modification data. The research is described in Baker et al (2015) Chromatin state analysis of the barley epigenome reveals a higher order structure defined by H3K27me1 and H3K27me3 abundance Plant J. 84:111-124. Chromosome sequences were manually assembled by incorporating PopSeq and IBGSC genetic map data to order as many Morex V3 contigs as possible. Gene and TE annotations from the IBGSC were then related to the new pseudo genome locations and are also available as tracks in the epigenome browser. To search for a gene of interest in the browser use the search bar next to the chromosome drop-down menu. Chromatin states were defined with ChromHMM and mapped to the genome. Coordinate files detailing the position of contigs, genes and TEs can be downloaded as gzipped files in the links at the bottom of this page. Histone modification peaks were called against the pseudo genome, but locations have been related back to individual contigs and are available to download below.
Access the barley epigenome browser here:
[sc:WebTable url=”https://ics.hutton.ac.uk/jbrowse/barley-chip/” urltext=”https://ics.hutton.ac.uk/jbrowse/barley-chip/” developers=”Katie Baker and Iain Milne”]Data for manually assembled barley genome (pseudo genome) and epigenome
The following are peak coordinates and fold changes for each histone modification in relation to the Morex V3 contigs:
The following are coordinate files for the positioning of contigs and associated gene and TE annotations in the barley pseudo genome.
Contigs:
Genes and TEs:
- pseudoGenome_chromosome_1.gff.gz
- pseudoGenome_chromosome_2.gff.gz
- pseudoGenome_chromosome_3.gff.gz
- pseudoGenome_chromosome_4.gff.gz
- pseudoGenome_chromosome_5.gff.gz
- pseudoGenome_chromosome_6.gff.gz
- pseudoGenome_chromosome_7.gff.gz