Probe CUST_9485_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
All Microarray Probes Designed to Gene DMG400002573
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
Microarray Signals from CUST_9485_PI426222305
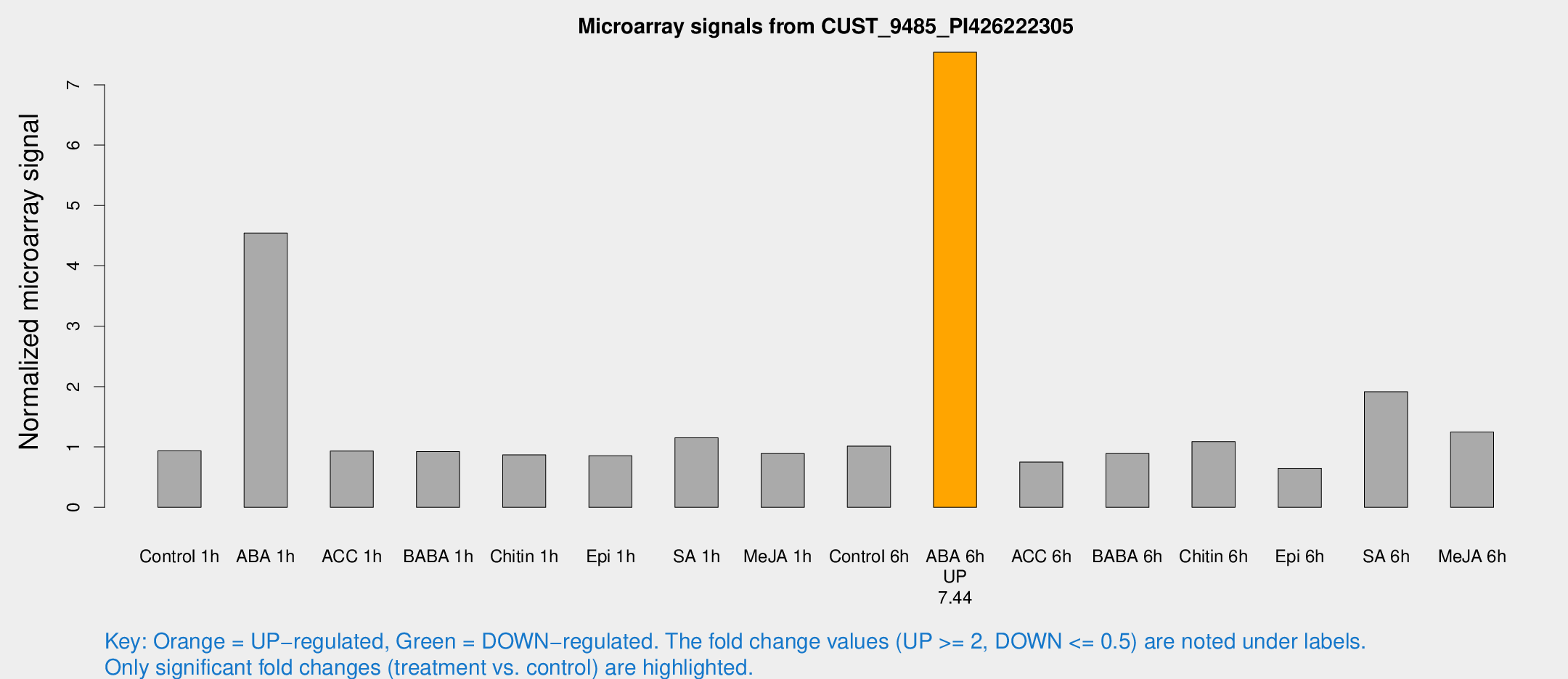
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 32.9296 | 3.70478 | 0.935709 | 0.128635 |
| ABA 1h | 146.016 | 26.8014 | 4.54437 | 0.754245 |
| ACC 1h | 35.934 | 9.07945 | 0.932072 | 0.198847 |
| BABA 1h | 34.2533 | 9.84364 | 0.922035 | 0.215836 |
| Chitin 1h | 27.6286 | 3.55178 | 0.86829 | 0.112477 |
| Epi 1h | 29.2768 | 10.4476 | 0.854684 | 0.344859 |
| SA 1h | 42.522 | 6.70639 | 1.15114 | 0.153141 |
| Me-JA 1h | 26.2175 | 4.67941 | 0.891136 | 0.126774 |
| Control 6h | 37.0825 | 7.35199 | 1.01364 | 0.270652 |
| ABA 6h | 290.481 | 50.7513 | 7.53959 | 1.10782 |
| ACC 6h | 31.2617 | 5.55199 | 0.750415 | 0.149444 |
| BABA 6h | 35.6546 | 5.34945 | 0.891533 | 0.123381 |
| Chitin 6h | 41.6023 | 6.7703 | 1.08919 | 0.200065 |
| Epi 6h | 27.2015 | 6.40534 | 0.648395 | 0.138867 |
| SA 6h | 69.2935 | 13.7574 | 1.91488 | 0.649108 |
| Me-JA 6h | 48.4314 | 16.0511 | 1.24777 | 0.319086 |
Source Transcript PGSC0003DMT400006609 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72770.1 | +3 | 4e-155 | 473 | 272/551 (49%) | HYPERSENSITIVE TO ABA1 | chr1:27390998-27392851 FORWARD LENGTH=511 |
