Probe CUST_9186_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
All Microarray Probes Designed to Gene DMG400002573
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
Microarray Signals from CUST_9186_PI426222305
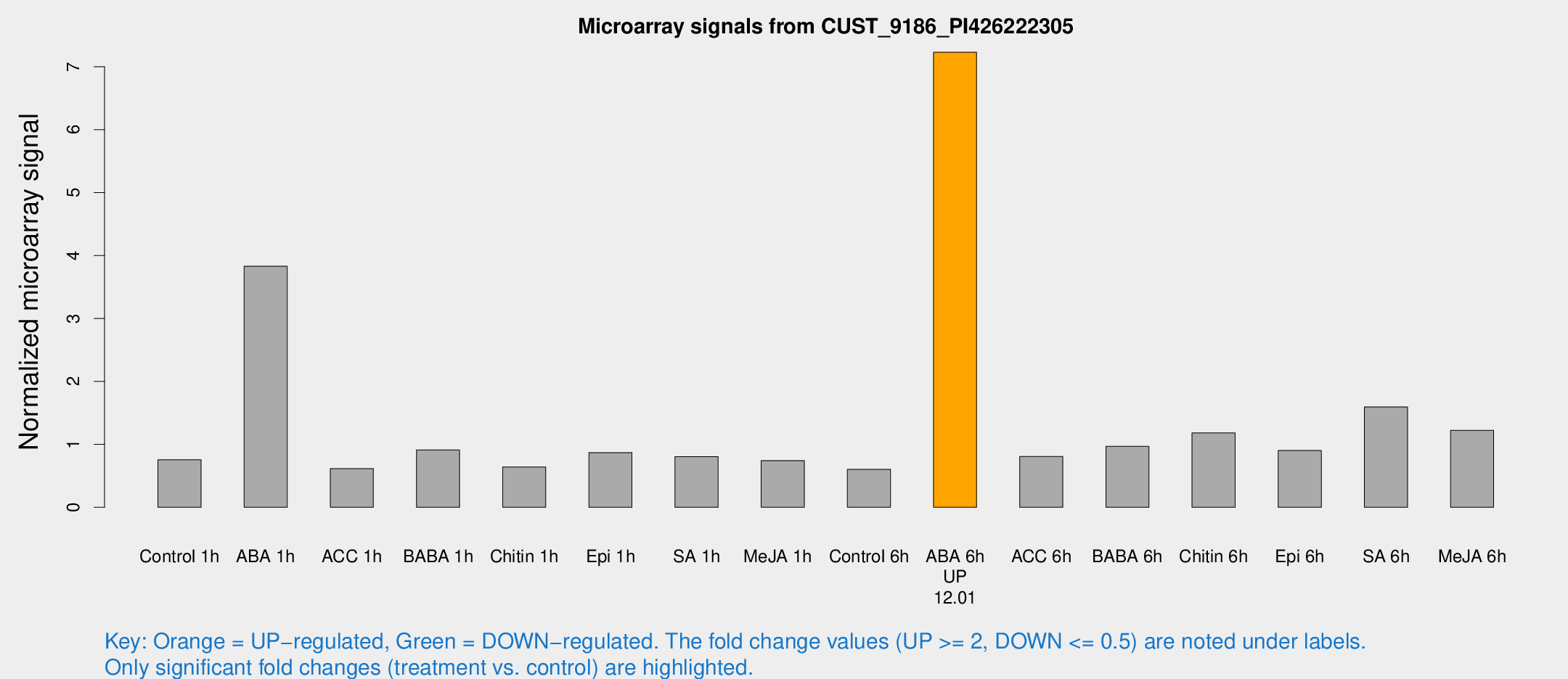
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 28.8135 | 3.79038 | 0.755208 | 0.0998502 |
| ABA 1h | 137.227 | 31.1262 | 3.83193 | 0.86196 |
| ACC 1h | 29.51 | 12.1542 | 0.614796 | 0.290203 |
| BABA 1h | 39.0099 | 13.134 | 0.91222 | 0.31783 |
| Chitin 1h | 24.0515 | 6.84461 | 0.641441 | 0.204761 |
| Epi 1h | 30.711 | 8.63058 | 0.869637 | 0.245617 |
| SA 1h | 33.0422 | 6.75606 | 0.806048 | 0.143377 |
| Me-JA 1h | 23.6433 | 3.82974 | 0.74212 | 0.166489 |
| Control 6h | 26.3209 | 9.66165 | 0.602304 | 0.200549 |
| ABA 6h | 301.938 | 51.4402 | 7.23125 | 1.05168 |
| ACC 6h | 36.1847 | 5.80916 | 0.808131 | 0.255677 |
| BABA 6h | 43.3712 | 10.1009 | 0.968075 | 0.207696 |
| Chitin 6h | 47.894 | 4.80183 | 1.18221 | 0.119059 |
| Epi 6h | 38.6689 | 4.76107 | 0.902267 | 0.109958 |
| SA 6h | 61.7556 | 10.0118 | 1.59488 | 0.441266 |
| Me-JA 6h | 47.6995 | 8.30272 | 1.2235 | 0.121464 |
Source Transcript PGSC0003DMT400006607 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72770.1 | +1 | 3e-52 | 189 | 85/113 (75%) | HYPERSENSITIVE TO ABA1 | chr1:27390998-27392851 FORWARD LENGTH=511 |
