Probe CUST_9304_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
All Microarray Probes Designed to Gene DMG400002573
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
Microarray Signals from CUST_9304_PI426222305
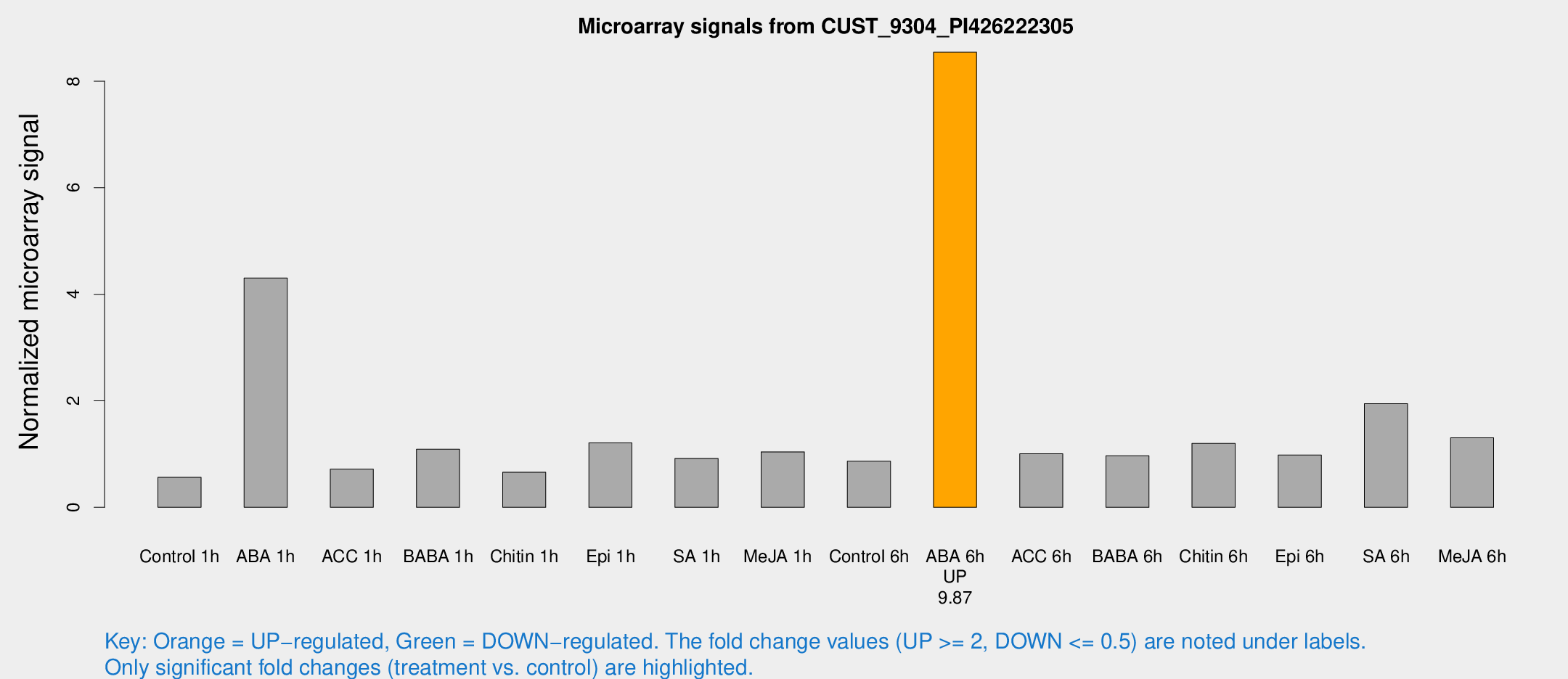
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.8569 | 4.43708 | 0.562717 | 0.196582 |
| ABA 1h | 142.425 | 42.0004 | 4.30584 | 1.4988 |
| ACC 1h | 27.6019 | 8.5549 | 0.715916 | 0.229453 |
| BABA 1h | 38.0626 | 10.4257 | 1.0903 | 0.234216 |
| Chitin 1h | 19.733 | 3.49114 | 0.657844 | 0.115894 |
| Epi 1h | 35.907 | 6.71128 | 1.21013 | 0.229738 |
| SA 1h | 33.6303 | 8.17014 | 0.91759 | 0.227248 |
| Me-JA 1h | 29.5061 | 6.03884 | 1.04117 | 0.288031 |
| Control 6h | 29.8688 | 5.73767 | 0.8658 | 0.124122 |
| ABA 6h | 311.8 | 53.5803 | 8.5441 | 1.18705 |
| ACC 6h | 38.5315 | 4.67892 | 1.00454 | 0.119394 |
| BABA 6h | 36.4952 | 4.41661 | 0.967518 | 0.118781 |
| Chitin 6h | 44.0592 | 9.09378 | 1.19908 | 0.273195 |
| Epi 6h | 36.9296 | 4.47002 | 0.980535 | 0.118108 |
| SA 6h | 68.3023 | 17.6958 | 1.94586 | 0.558061 |
| Me-JA 6h | 44.4021 | 7.52155 | 1.30462 | 0.208877 |
Source Transcript PGSC0003DMT400006608 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72770.1 | +2 | 2e-170 | 521 | 291/542 (54%) | HYPERSENSITIVE TO ABA1 | chr1:27390998-27392851 FORWARD LENGTH=511 |
