Probe CUST_9513_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
All Microarray Probes Designed to Gene DMG400002573
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
Microarray Signals from CUST_9513_PI426222305
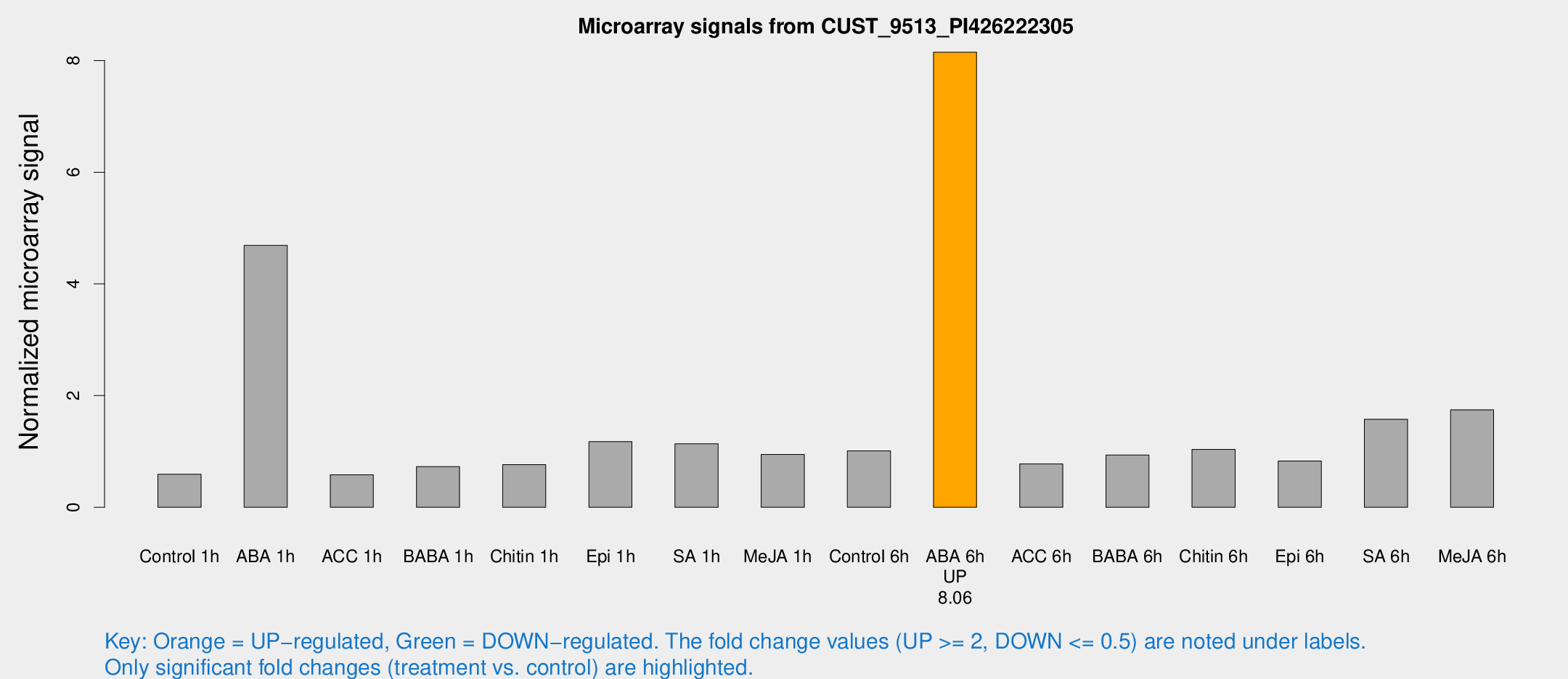
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.1822 | 6.88129 | 0.592841 | 0.27334 |
| ABA 1h | 145.795 | 34.6603 | 4.69066 | 1.12284 |
| ACC 1h | 24.934 | 9.26398 | 0.580842 | 0.332974 |
| BABA 1h | 25.2195 | 6.53469 | 0.729647 | 0.159302 |
| Chitin 1h | 23.1885 | 3.59594 | 0.764439 | 0.141399 |
| Epi 1h | 33.6812 | 3.82533 | 1.1752 | 0.137498 |
| SA 1h | 40.9002 | 9.28655 | 1.13635 | 0.256293 |
| Me-JA 1h | 27.0252 | 6.9582 | 0.946567 | 0.157994 |
| Control 6h | 34.3746 | 6.39151 | 1.01105 | 0.143069 |
| ABA 6h | 293.473 | 48.9454 | 8.14798 | 1.10783 |
| ACC 6h | 29.412 | 4.49659 | 0.776891 | 0.115369 |
| BABA 6h | 37.6201 | 10.0466 | 0.935811 | 0.281046 |
| Chitin 6h | 38.6982 | 10.192 | 1.03734 | 0.311644 |
| Epi 6h | 33.0174 | 7.95793 | 0.829958 | 0.179301 |
| SA 6h | 55.8441 | 14.3368 | 1.57563 | 0.69188 |
| Me-JA 6h | 58.5398 | 9.37976 | 1.74475 | 0.146605 |
Source Transcript PGSC0003DMT400006611 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72770.1 | +2 | 2e-161 | 498 | 291/590 (49%) | HYPERSENSITIVE TO ABA1 | chr1:27390998-27392851 FORWARD LENGTH=511 |
