Probe CUST_9294_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
All Microarray Probes Designed to Gene DMG400002573
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9304_PI426222305 | JHI_St_60k_v1 | DMT400006608 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9513_PI426222305 | JHI_St_60k_v1 | DMT400006611 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9294_PI426222305 | JHI_St_60k_v1 | DMT400006610 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9186_PI426222305 | JHI_St_60k_v1 | DMT400006607 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
| CUST_9485_PI426222305 | JHI_St_60k_v1 | DMT400006609 | GCAGATCAAAGATCTTACTTATCCTGGACAAGTATATTGAGATTTCGAAGAAAGAGTAGA |
Microarray Signals from CUST_9294_PI426222305
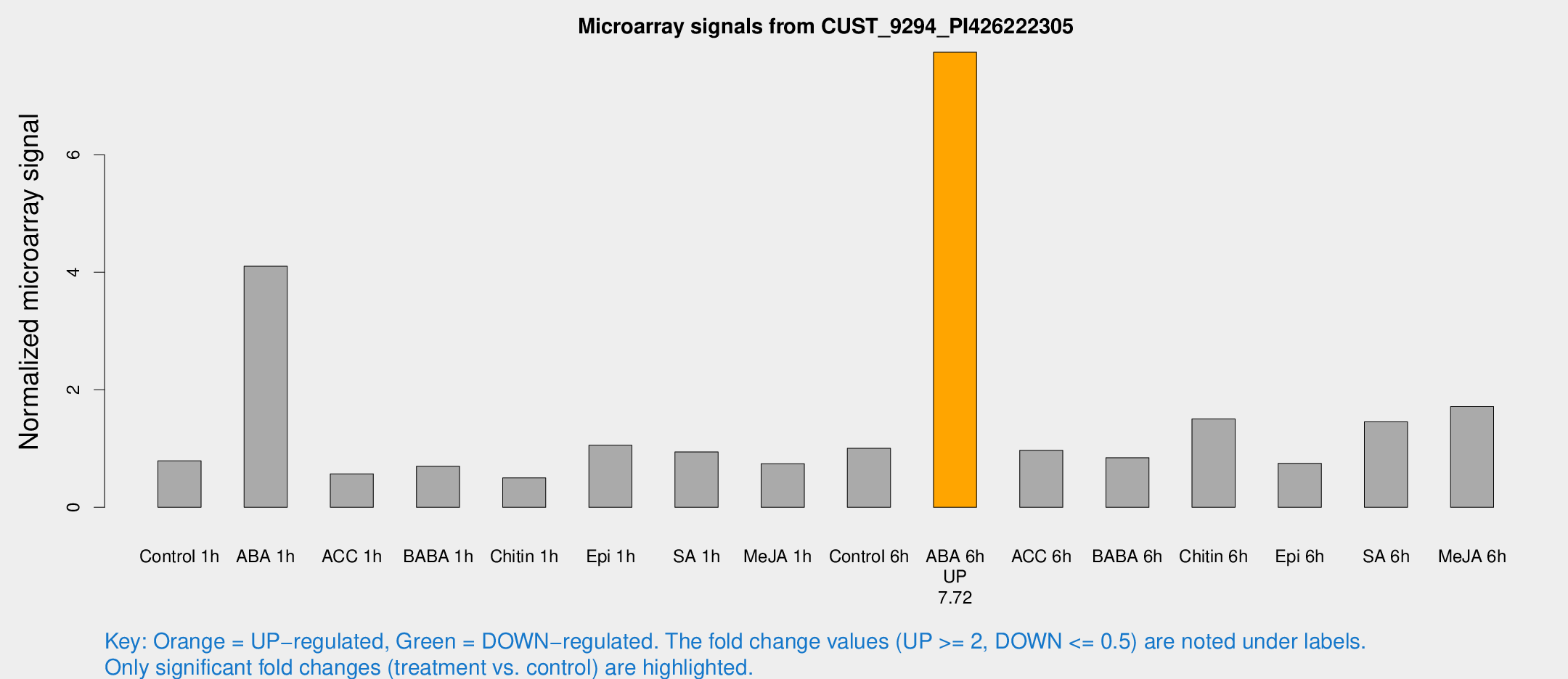
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 28.4186 | 4.16782 | 0.790059 | 0.0988613 |
| ABA 1h | 134.956 | 29.8187 | 4.10293 | 0.87968 |
| ACC 1h | 27.4541 | 10.7981 | 0.567842 | 0.393487 |
| BABA 1h | 25.3015 | 6.40777 | 0.700144 | 0.124522 |
| Chitin 1h | 20.002 | 7.39886 | 0.500025 | 0.359556 |
| Epi 1h | 34.3342 | 8.12268 | 1.05536 | 0.291491 |
| SA 1h | 34.5496 | 4.28654 | 0.941631 | 0.105083 |
| Me-JA 1h | 22.3078 | 4.50711 | 0.741855 | 0.201972 |
| Control 6h | 37.7454 | 10.0244 | 1.00293 | 0.214576 |
| ABA 6h | 301.988 | 57.8882 | 7.74579 | 1.32712 |
| ACC 6h | 39.3078 | 4.45144 | 0.968711 | 0.142054 |
| BABA 6h | 34.7123 | 7.48793 | 0.844713 | 0.18986 |
| Chitin 6h | 57.2286 | 7.98618 | 1.50305 | 0.241744 |
| Epi 6h | 29.8454 | 4.01033 | 0.748023 | 0.0996606 |
| SA 6h | 51.8188 | 8.35532 | 1.45632 | 0.18289 |
| Me-JA 6h | 62.5821 | 13.6551 | 1.71362 | 0.25816 |
Source Transcript PGSC0003DMT400006610 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72770.1 | +3 | 2e-164 | 496 | 272/503 (54%) | HYPERSENSITIVE TO ABA1 | chr1:27390998-27392851 FORWARD LENGTH=511 |
