Probe CUST_9058_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
All Microarray Probes Designed to Gene DMG401018664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
Microarray Signals from CUST_9058_PI426222305
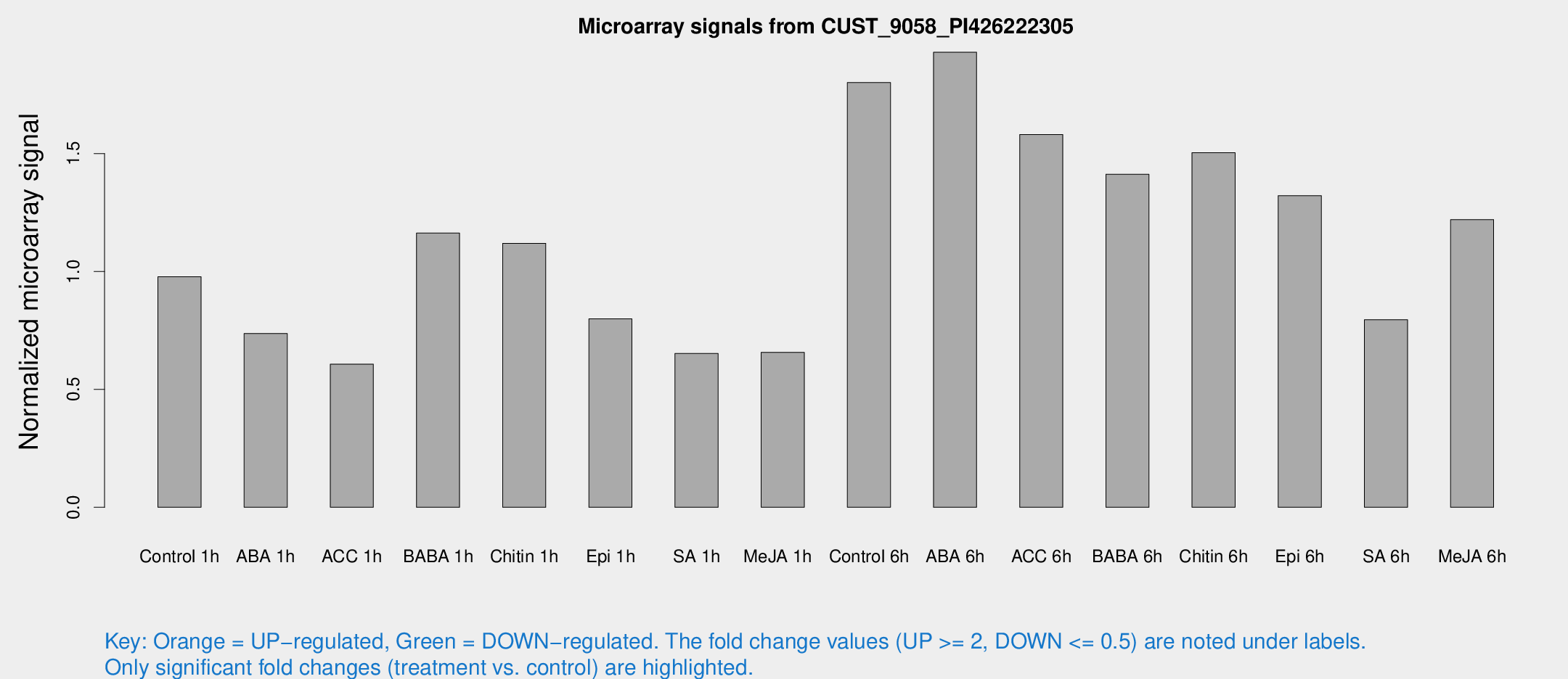
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 52.1696 | 4.71328 | 0.977977 | 0.0802491 |
| ABA 1h | 35.3241 | 5.41228 | 0.737261 | 0.10438 |
| ACC 1h | 36.3034 | 10.6716 | 0.606785 | 0.161736 |
| BABA 1h | 60.6032 | 8.742 | 1.16291 | 0.0937133 |
| Chitin 1h | 55.8567 | 12.5509 | 1.11956 | 0.240049 |
| Epi 1h | 37.0362 | 3.8993 | 0.799098 | 0.0811997 |
| SA 1h | 36.7551 | 6.95584 | 0.65262 | 0.112302 |
| Me-JA 1h | 29.9274 | 6.68284 | 0.657273 | 0.0975426 |
| Control 6h | 106.36 | 29.7453 | 1.8013 | 0.491066 |
| ABA 6h | 118.093 | 29.8029 | 1.92992 | 0.536908 |
| ACC 6h | 106.404 | 33.0919 | 1.5811 | 0.337891 |
| BABA 6h | 88.6115 | 19.0612 | 1.4125 | 0.313749 |
| Chitin 6h | 86.4615 | 11.8612 | 1.50363 | 0.218849 |
| Epi 6h | 83.7639 | 18.9908 | 1.32143 | 0.442276 |
| SA 6h | 57.7304 | 29.0657 | 0.795773 | 0.874991 |
| Me-JA 6h | 72.918 | 25.3472 | 1.21991 | 0.354621 |
Source Transcript PGSC0003DMT400048033 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37020.1 | +2 | 2e-114 | 354 | 166/191 (87%) | auxin response factor 8 | chr5:14630151-14634106 FORWARD LENGTH=811 |
