Probe CUST_9031_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
All Microarray Probes Designed to Gene DMG401018664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
Microarray Signals from CUST_9031_PI426222305
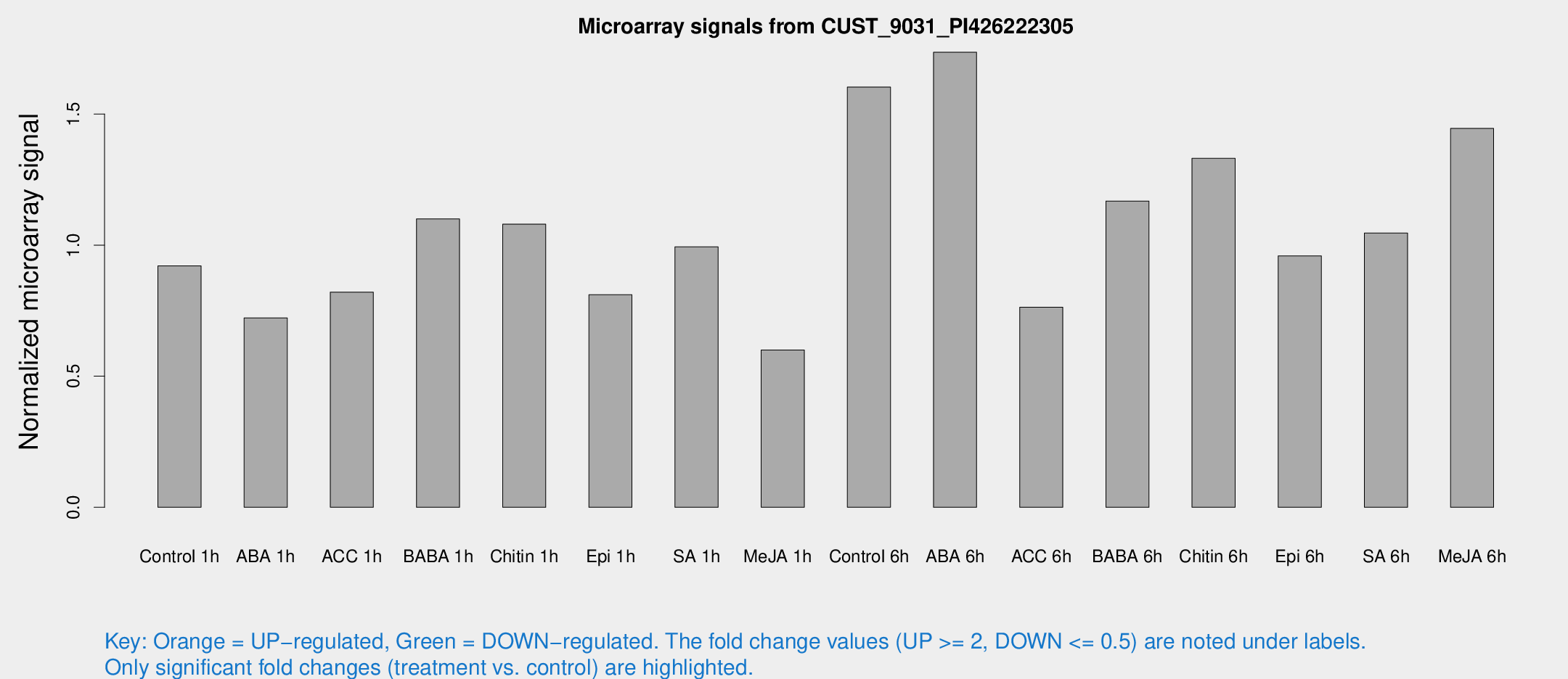
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 79.4918 | 13.6252 | 0.920914 | 0.103617 |
| ABA 1h | 53.7219 | 4.59172 | 0.722559 | 0.0618497 |
| ACC 1h | 73.2395 | 12.5558 | 0.82088 | 0.0954146 |
| BABA 1h | 91.3288 | 13.7652 | 1.09995 | 0.0789864 |
| Chitin 1h | 82.4342 | 8.23927 | 1.07993 | 0.0855483 |
| Epi 1h | 59.5436 | 6.15554 | 0.810682 | 0.0914334 |
| SA 1h | 85.8854 | 6.0494 | 0.993317 | 0.0938064 |
| Me-JA 1h | 44.2427 | 10.5876 | 0.599627 | 0.185835 |
| Control 6h | 139.511 | 24.2238 | 1.60261 | 0.174312 |
| ABA 6h | 161.192 | 29.9343 | 1.73574 | 0.267813 |
| ACC 6h | 73.5627 | 6.13202 | 0.763244 | 0.107626 |
| BABA 6h | 116.077 | 26.4148 | 1.16808 | 0.255235 |
| Chitin 6h | 120.443 | 13.5137 | 1.33076 | 0.104079 |
| Epi 6h | 103.202 | 33.8225 | 0.958641 | 0.462525 |
| SA 6h | 91.3826 | 18.7912 | 1.04594 | 0.328154 |
| Me-JA 6h | 131.153 | 33.9541 | 1.44504 | 0.321616 |
Source Transcript PGSC0003DMT400048035 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37020.1 | +3 | 0.0 | 632 | 370/615 (60%) | auxin response factor 8 | chr5:14630151-14634106 FORWARD LENGTH=811 |
