Probe CUST_8922_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
All Microarray Probes Designed to Gene DMG401018664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
Microarray Signals from CUST_8922_PI426222305
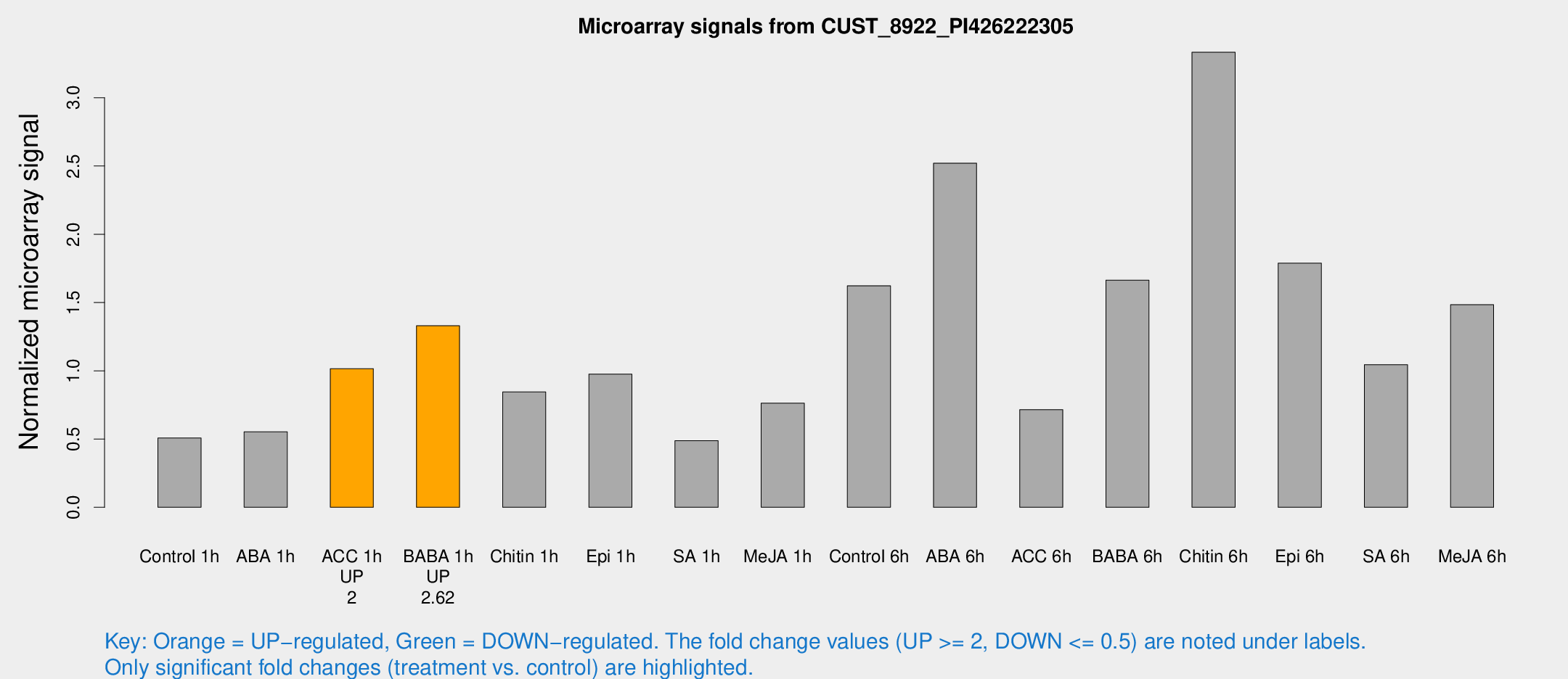
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.47242 | 3.76019 | 0.507393 | 0.294061 |
| ABA 1h | 6.24363 | 3.6361 | 0.552444 | 0.320407 |
| ACC 1h | 13.3277 | 4.99769 | 1.01535 | 0.389008 |
| BABA 1h | 17.2398 | 4.02535 | 1.32954 | 0.355546 |
| Chitin 1h | 11.6272 | 5.2111 | 0.844901 | 0.399585 |
| Epi 1h | 11.2018 | 3.70388 | 0.975805 | 0.351961 |
| SA 1h | 6.37808 | 3.69709 | 0.487816 | 0.282748 |
| Me-JA 1h | 7.97466 | 3.84319 | 0.7628 | 0.373471 |
| Control 6h | 26.6866 | 10.1226 | 1.6229 | 0.925422 |
| ABA 6h | 38.8644 | 12.3109 | 2.52072 | 0.9133 |
| ACC 6h | 10.6611 | 4.79396 | 0.715381 | 0.329024 |
| BABA 6h | 30.8987 | 11.941 | 1.66387 | 1.16478 |
| Chitin 6h | 69.7831 | 42.4425 | 3.33349 | 3.20847 |
| Epi 6h | 28.7544 | 9.29779 | 1.78839 | 0.77133 |
| SA 6h | 20.6221 | 13.7396 | 1.04524 | 1.33385 |
| Me-JA 6h | 22.123 | 8.08892 | 1.48372 | 0.547979 |
Source Transcript PGSC0003DMT400048031 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G30330.2 | +1 | 3e-120 | 367 | 186/203 (92%) | auxin response factor 6 | chr1:10686125-10690036 REVERSE LENGTH=935 |
