Probe CUST_8917_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
All Microarray Probes Designed to Gene DMG401018664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
Microarray Signals from CUST_8917_PI426222305
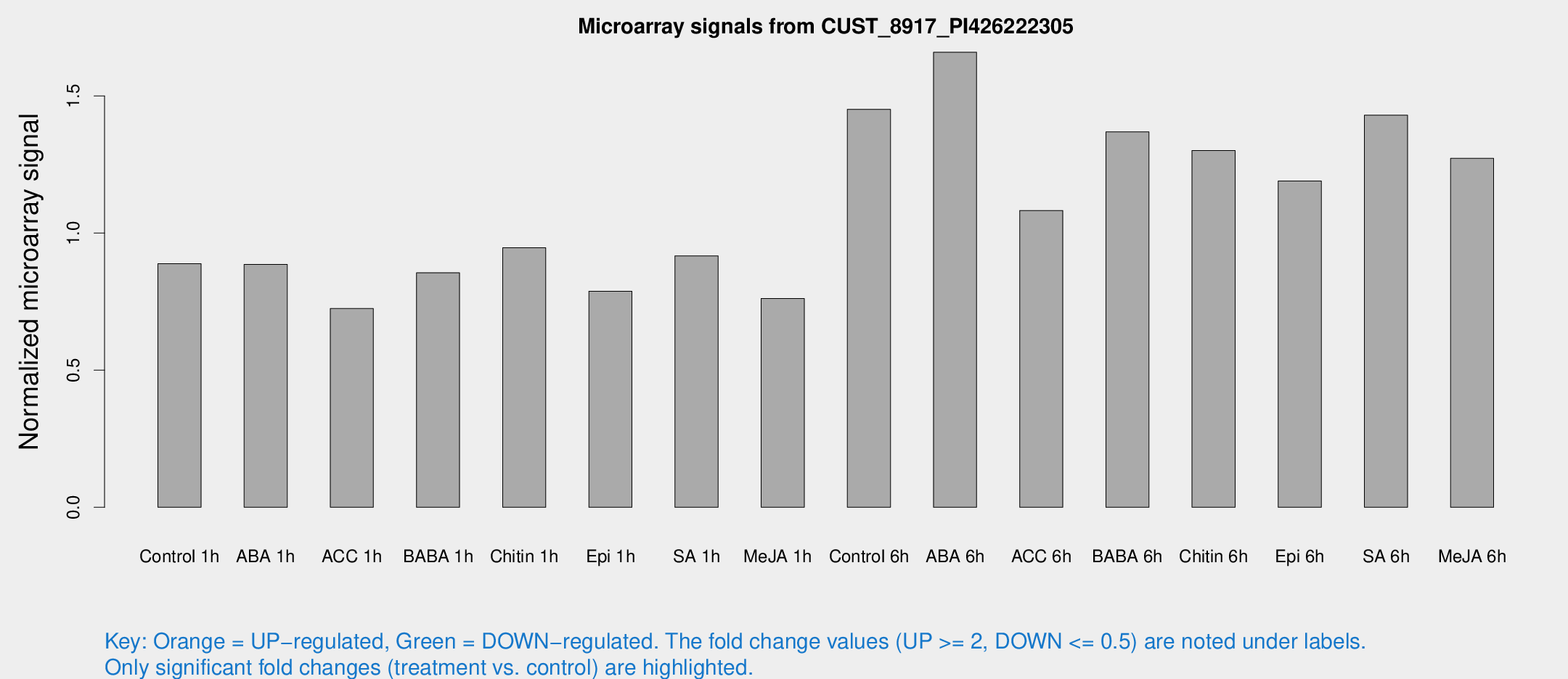
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 91.9727 | 18.1587 | 0.888142 | 0.114387 |
| ABA 1h | 78.5511 | 5.63692 | 0.885963 | 0.0626164 |
| ACC 1h | 79.6739 | 18.7414 | 0.724779 | 0.151559 |
| BABA 1h | 84.0132 | 12.4942 | 0.85528 | 0.0712553 |
| Chitin 1h | 86.8244 | 13.4619 | 0.946429 | 0.0702746 |
| Epi 1h | 69.0943 | 8.70737 | 0.787999 | 0.0996955 |
| SA 1h | 94.4119 | 7.0847 | 0.91665 | 0.0645088 |
| Me-JA 1h | 63.1894 | 8.72546 | 0.761749 | 0.060667 |
| Control 6h | 159.91 | 42.9769 | 1.45156 | 0.360925 |
| ABA 6h | 181.724 | 33.5595 | 1.65958 | 0.242014 |
| ACC 6h | 128.141 | 22.0695 | 1.08253 | 0.145785 |
| BABA 6h | 154.748 | 16.6675 | 1.3691 | 0.105536 |
| Chitin 6h | 138.744 | 9.55903 | 1.3009 | 0.110933 |
| Epi 6h | 137.44 | 21.8151 | 1.18997 | 0.27059 |
| SA 6h | 143.91 | 19.3805 | 1.4299 | 0.0908122 |
| Me-JA 6h | 138.492 | 36.2835 | 1.27241 | 0.327239 |
Source Transcript PGSC0003DMT400048036 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37020.1 | +3 | 0.0 | 671 | 389/642 (61%) | auxin response factor 8 | chr5:14630151-14634106 FORWARD LENGTH=811 |
