Probe CUST_9134_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
All Microarray Probes Designed to Gene DMG401018664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8922_PI426222305 | JHI_St_60k_v1 | DMT400048031 | TTGCGTACTCAATGTTATGCTGTTAGGAAAGTGAAGCTGTACAATCCTGAGGTTTCCTTG |
| CUST_9058_PI426222305 | JHI_St_60k_v1 | DMT400048033 | TTTACGCCGTGTTAAGTGTATTTGGCTTTAATCTAACAGCTGGTGGTTCTTTAAGTTTGC |
| CUST_8917_PI426222305 | JHI_St_60k_v1 | DMT400048036 | GAGTTGTAAACTTGTTAATGCAGCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGG |
| CUST_9031_PI426222305 | JHI_St_60k_v1 | DMT400048035 | GGGATTACACTTGGTATGTTATTGGCTTTTTACCATCTTTTGTTCAAACCAAGACATGAT |
| CUST_9134_PI426222305 | JHI_St_60k_v1 | DMT400048034 | GCTAAACTTTTGCGAGTAAATTCCCTTTTTCGAAAAGGTTTCTCCAGAACCATGTCCGCG |
Microarray Signals from CUST_9134_PI426222305
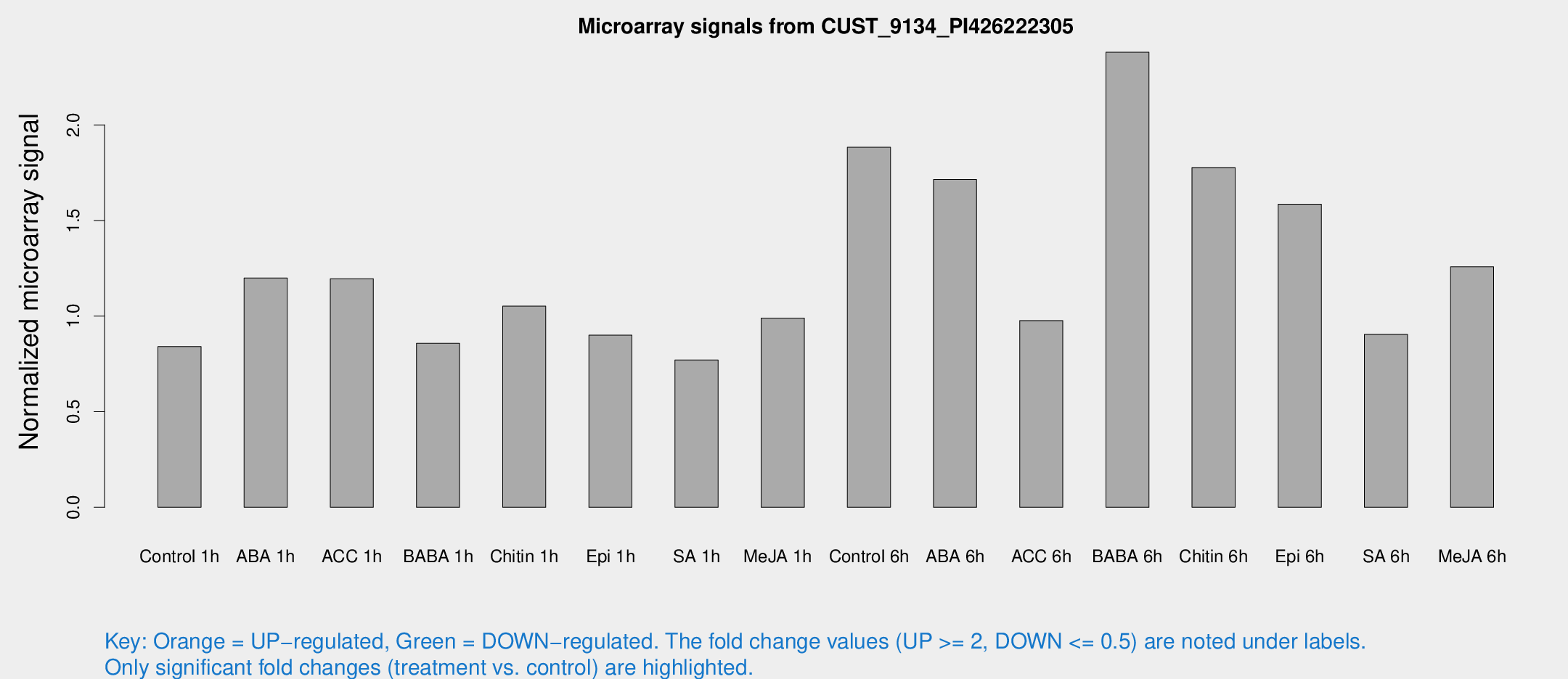
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.7672 | 3.14362 | 0.840673 | 0.459923 |
| ABA 1h | 7.41659 | 3.05049 | 1.19911 | 0.520512 |
| ACC 1h | 8.96581 | 3.74148 | 1.1955 | 0.570157 |
| BABA 1h | 5.67044 | 3.28882 | 0.857568 | 0.496728 |
| Chitin 1h | 6.65469 | 3.14172 | 1.05185 | 0.523069 |
| Epi 1h | 5.30856 | 3.08361 | 0.900201 | 0.518268 |
| SA 1h | 5.42358 | 3.14628 | 0.770227 | 0.446607 |
| Me-JA 1h | 5.54989 | 3.23275 | 0.989342 | 0.574362 |
| Control 6h | 14.9657 | 4.69156 | 1.88343 | 0.660292 |
| ABA 6h | 14.3787 | 4.96556 | 1.71411 | 0.811308 |
| ACC 6h | 7.95288 | 4.01077 | 0.976646 | 0.489207 |
| BABA 6h | 19.1565 | 3.8472 | 2.38018 | 0.517729 |
| Chitin 6h | 14.9024 | 4.76207 | 1.77707 | 0.827282 |
| Epi 6h | 13.2008 | 3.83941 | 1.58522 | 0.574333 |
| SA 6h | 6.16376 | 3.38524 | 0.904489 | 0.500073 |
| Me-JA 6h | 9.46223 | 3.26728 | 1.25776 | 0.525836 |
Source Transcript PGSC0003DMT400048034 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G37020.1 | +1 | 0.0 | 863 | 480/750 (64%) | auxin response factor 8 | chr5:14630151-14634106 FORWARD LENGTH=811 |
