Probe CUST_5311_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
All Microarray Probes Designed to Gene DMG400014902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
Microarray Signals from CUST_5311_PI426222305
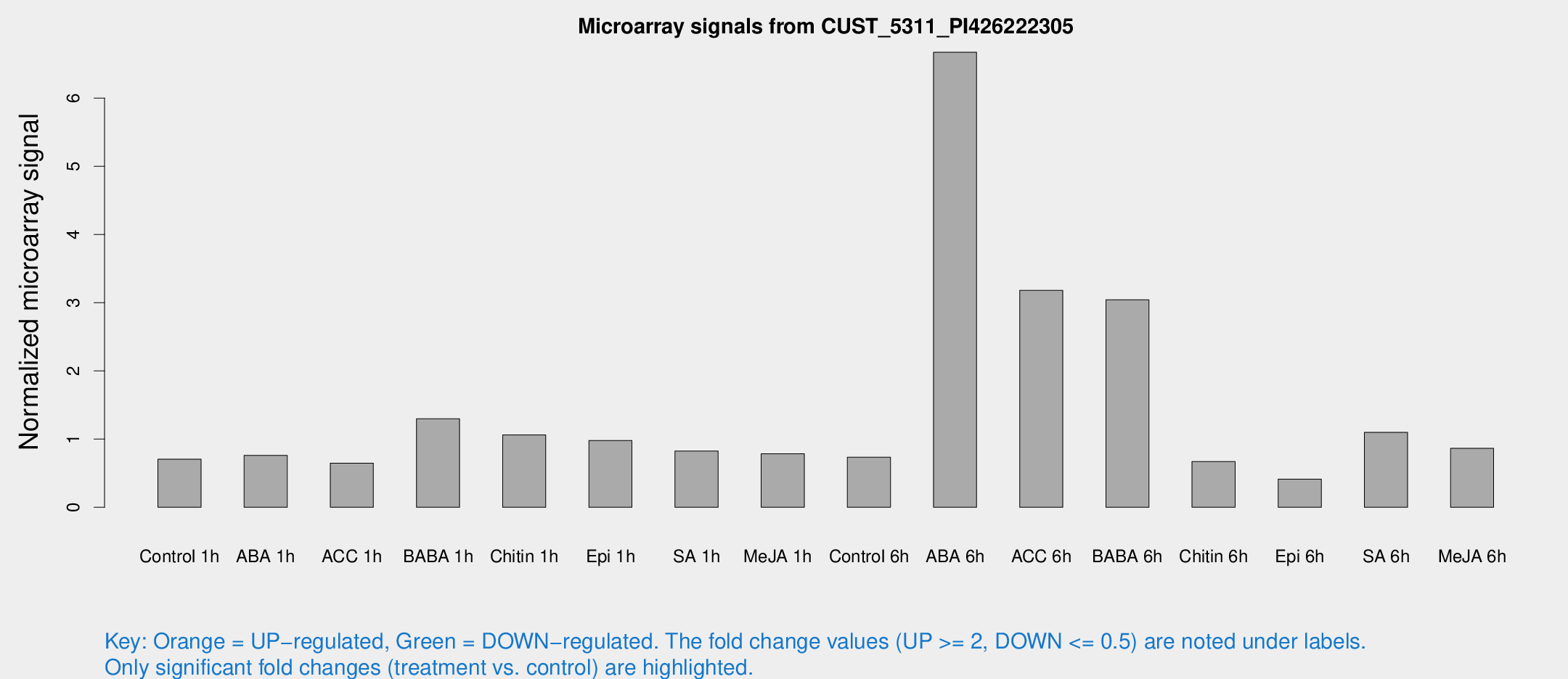
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.1526 | 3.33848 | 0.705283 | 0.131706 |
| ABA 1h | 22.451 | 8.63109 | 0.760296 | 0.517957 |
| ACC 1h | 20.9523 | 8.10338 | 0.647063 | 0.322472 |
| BABA 1h | 32.5507 | 4.63838 | 1.29742 | 0.270812 |
| Chitin 1h | 25.8938 | 5.84687 | 1.06289 | 0.289027 |
| Epi 1h | 22.6013 | 5.01262 | 0.980555 | 0.21917 |
| SA 1h | 22.6961 | 4.80116 | 0.824466 | 0.164499 |
| Me-JA 1h | 17.1996 | 3.98872 | 0.785165 | 0.167041 |
| Control 6h | 20.6475 | 5.51375 | 0.734542 | 0.182907 |
| ABA 6h | 201.13 | 58.8125 | 6.67224 | 2.09614 |
| ACC 6h | 99.3995 | 23.4622 | 3.1809 | 1.48158 |
| BABA 6h | 98.857 | 36.7189 | 3.04285 | 1.04756 |
| Chitin 6h | 22.8993 | 8.65398 | 0.670396 | 0.460064 |
| Epi 6h | 12.4419 | 3.9169 | 0.413417 | 0.152532 |
| SA 6h | 45.1967 | 24.391 | 1.09782 | 2.04187 |
| Me-JA 6h | 30.8484 | 16.1102 | 0.864425 | 0.691029 |
Source Transcript PGSC0003DMT400038580 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50660.1 | +1 | 0.0 | 669 | 343/482 (71%) | Cytochrome P450 superfamily protein | chr3:18814262-18817168 REVERSE LENGTH=513 |
