Probe CUST_5165_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
All Microarray Probes Designed to Gene DMG400014902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
Microarray Signals from CUST_5165_PI426222305
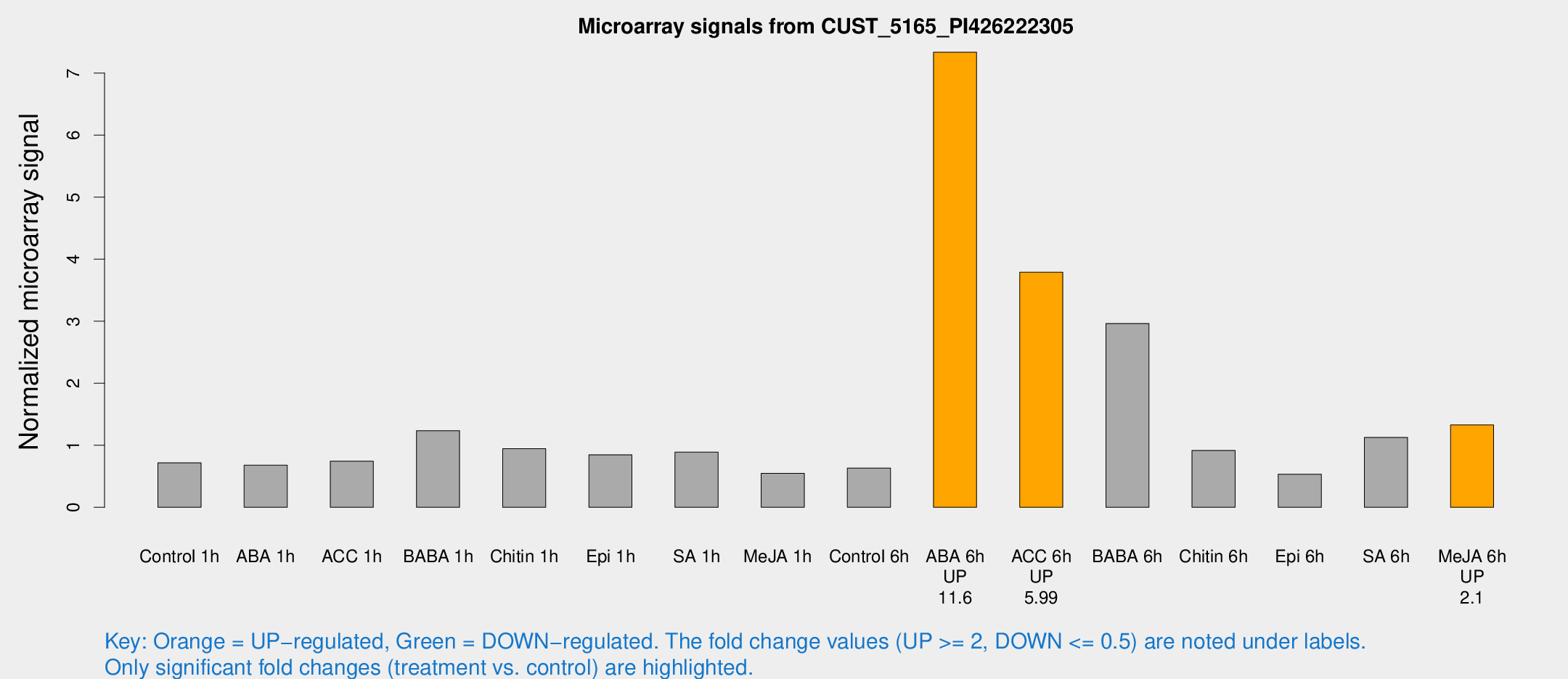
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.7887 | 3.19222 | 0.716148 | 0.124207 |
| ABA 1h | 17.4937 | 5.68189 | 0.677938 | 0.237086 |
| ACC 1h | 24.7578 | 9.61301 | 0.744311 | 0.386617 |
| BABA 1h | 33.7989 | 9.05917 | 1.23556 | 0.27454 |
| Chitin 1h | 22.4507 | 3.26024 | 0.944952 | 0.215242 |
| Epi 1h | 23.8229 | 8.70022 | 0.846519 | 0.545513 |
| SA 1h | 24.0714 | 3.43002 | 0.888618 | 0.133196 |
| Me-JA 1h | 15.1983 | 7.77704 | 0.547863 | 0.313501 |
| Control 6h | 17.1837 | 3.44712 | 0.632712 | 0.134363 |
| ABA 6h | 222.667 | 63.0808 | 7.33745 | 2.11075 |
| ACC 6h | 114.368 | 13.3129 | 3.78984 | 1.04125 |
| BABA 6h | 104.937 | 48.8317 | 2.9644 | 1.33523 |
| Chitin 6h | 25.5076 | 3.70731 | 0.916779 | 0.135319 |
| Epi 6h | 15.994 | 3.68657 | 0.533369 | 0.134217 |
| SA 6h | 46.0382 | 23.2524 | 1.12604 | 2.01382 |
| Me-JA 6h | 36.9015 | 10.465 | 1.32788 | 0.261356 |
Source Transcript PGSC0003DMT400038582 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50660.1 | +2 | 0.0 | 520 | 262/365 (72%) | Cytochrome P450 superfamily protein | chr3:18814262-18817168 REVERSE LENGTH=513 |
