Probe CUST_5104_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
All Microarray Probes Designed to Gene DMG400014902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
Microarray Signals from CUST_5104_PI426222305
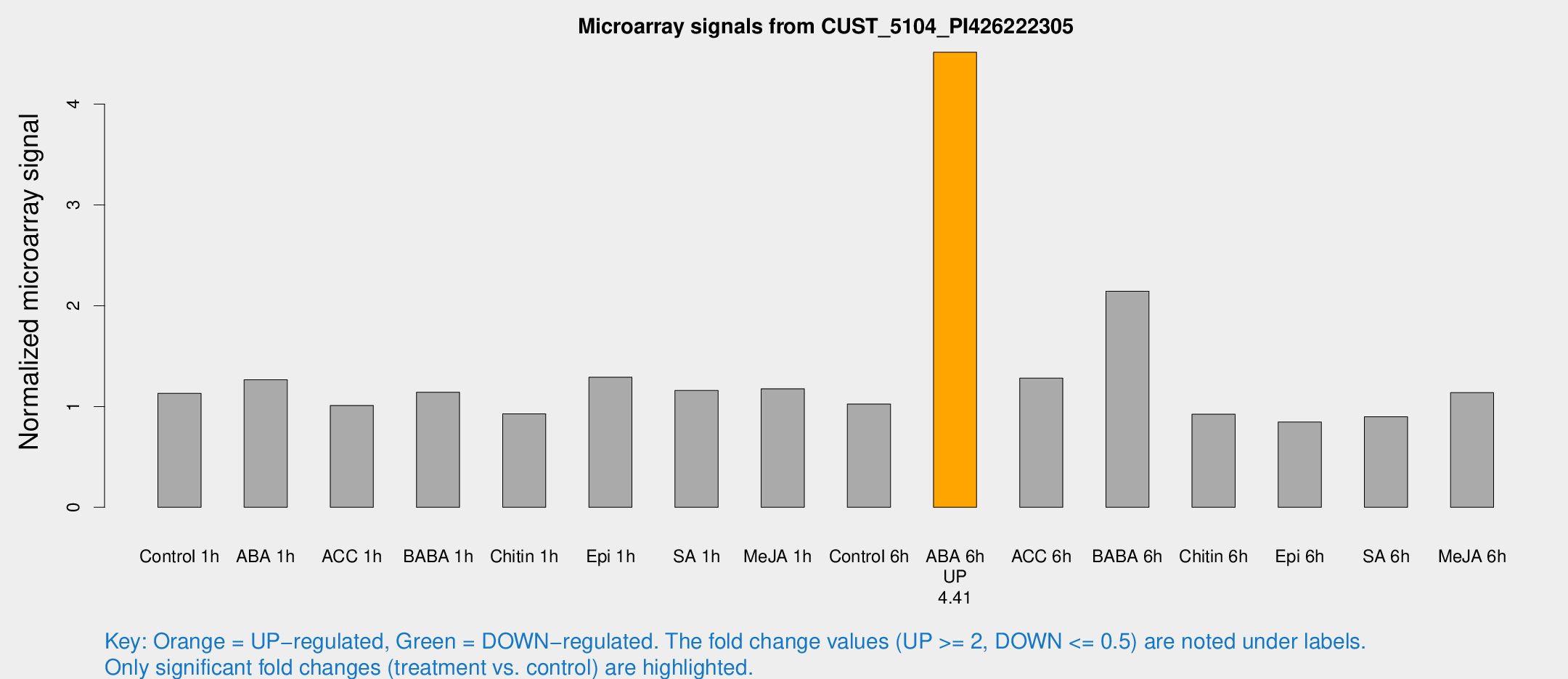
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.29131 | 3.1792 | 1.13089 | 0.535782 |
| ABA 1h | 8.3124 | 3.0959 | 1.26605 | 0.592102 |
| ACC 1h | 6.85334 | 3.57374 | 1.01036 | 0.532871 |
| BABA 1h | 7.51737 | 3.35393 | 1.14198 | 0.559746 |
| Chitin 1h | 5.36894 | 3.1275 | 0.927551 | 0.525974 |
| Epi 1h | 8.16122 | 3.11533 | 1.28977 | 0.601433 |
| SA 1h | 8.42866 | 3.14536 | 1.15881 | 0.509329 |
| Me-JA 1h | 6.37882 | 3.19342 | 1.17584 | 0.604604 |
| Control 6h | 6.85262 | 3.2523 | 1.02355 | 0.501732 |
| ABA 6h | 31.6542 | 3.80222 | 4.51479 | 0.546019 |
| ACC 6h | 10.9699 | 4.01575 | 1.28079 | 0.619904 |
| BABA 6h | 17.4067 | 5.58064 | 2.14402 | 0.698851 |
| Chitin 6h | 6.44828 | 3.54594 | 0.922797 | 0.509347 |
| Epi 6h | 6.31602 | 3.70144 | 0.84621 | 0.490679 |
| SA 6h | 5.82489 | 3.37609 | 0.899088 | 0.521075 |
| Me-JA 6h | 7.66964 | 3.18706 | 1.13729 | 0.507009 |
Source Transcript PGSC0003DMT400038577 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50660.1 | +3 | 4e-25 | 105 | 45/60 (75%) | Cytochrome P450 superfamily protein | chr3:18814262-18817168 REVERSE LENGTH=513 |
