Probe CUST_4985_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
All Microarray Probes Designed to Gene DMG400014902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
Microarray Signals from CUST_4985_PI426222305
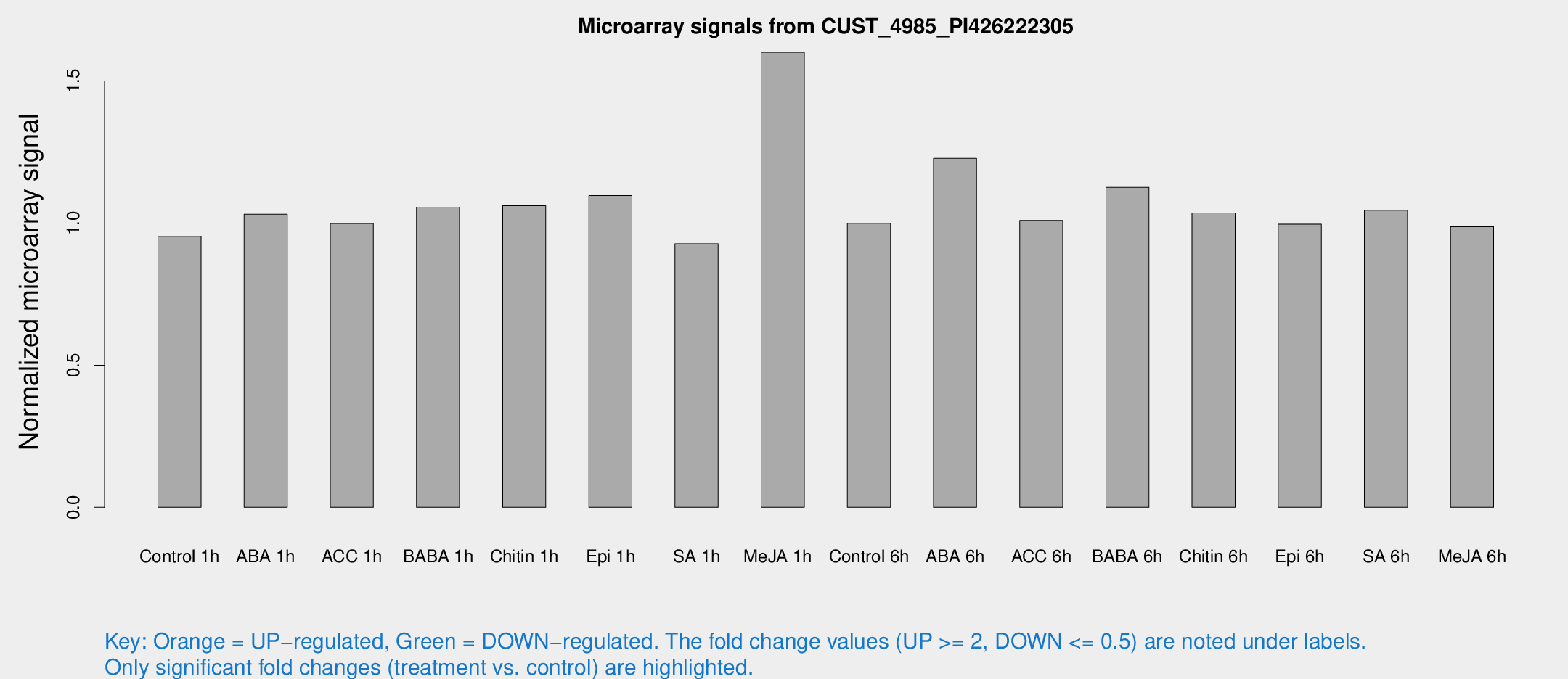
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.68604 | 3.29392 | 0.953317 | 0.552021 |
| ABA 1h | 5.44372 | 3.15562 | 1.03112 | 0.597112 |
| ACC 1h | 6.11895 | 3.54867 | 0.998593 | 0.578657 |
| BABA 1h | 6.07233 | 3.51753 | 1.05626 | 0.611631 |
| Chitin 1h | 5.7054 | 3.31329 | 1.06108 | 0.61446 |
| Epi 1h | 5.67366 | 3.29482 | 1.09669 | 0.63522 |
| SA 1h | 5.68228 | 3.29489 | 0.927514 | 0.537653 |
| Me-JA 1h | 8.34435 | 3.44147 | 1.601 | 0.766142 |
| Control 6h | 5.97032 | 3.45754 | 0.999082 | 0.578563 |
| ABA 6h | 8.37721 | 3.5904 | 1.22764 | 0.599138 |
| ACC 6h | 7.0765 | 4.20557 | 1.00942 | 0.584524 |
| BABA 6h | 7.58533 | 3.83862 | 1.12553 | 0.583653 |
| Chitin 6h | 6.57585 | 3.80911 | 1.0357 | 0.599695 |
| Epi 6h | 6.74407 | 3.93505 | 0.996223 | 0.577098 |
| SA 6h | 6.16718 | 3.57253 | 1.04539 | 0.605363 |
| Me-JA 6h | 5.86401 | 3.40354 | 0.987119 | 0.572576 |
Source Transcript PGSC0003DMT400038579 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50660.1 | +2 | 3e-143 | 425 | 225/332 (68%) | Cytochrome P450 superfamily protein | chr3:18814262-18817168 REVERSE LENGTH=513 |
