Probe CUST_5233_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
All Microarray Probes Designed to Gene DMG400014902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5165_PI426222305 | JHI_St_60k_v1 | DMT400038582 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_4985_PI426222305 | JHI_St_60k_v1 | DMT400038579 | CTGCTGTTCAACAACTAACAATTTCTACAGTTTACATTCAACTTTATGGCAGAGCATATC |
| CUST_5233_PI426222305 | JHI_St_60k_v1 | DMT400038581 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
| CUST_5104_PI426222305 | JHI_St_60k_v1 | DMT400038577 | GTATGTCCAGCGTACCAAACGACCCTTAGAACTATATATGTATATATGTGTATGGTAAAA |
| CUST_5311_PI426222305 | JHI_St_60k_v1 | DMT400038580 | GAAATCATCAATGTCTGACTTAGAGTTTTTTCTTTTTCTTGTTCCTCCAATCTTGGCAGT |
Microarray Signals from CUST_5233_PI426222305
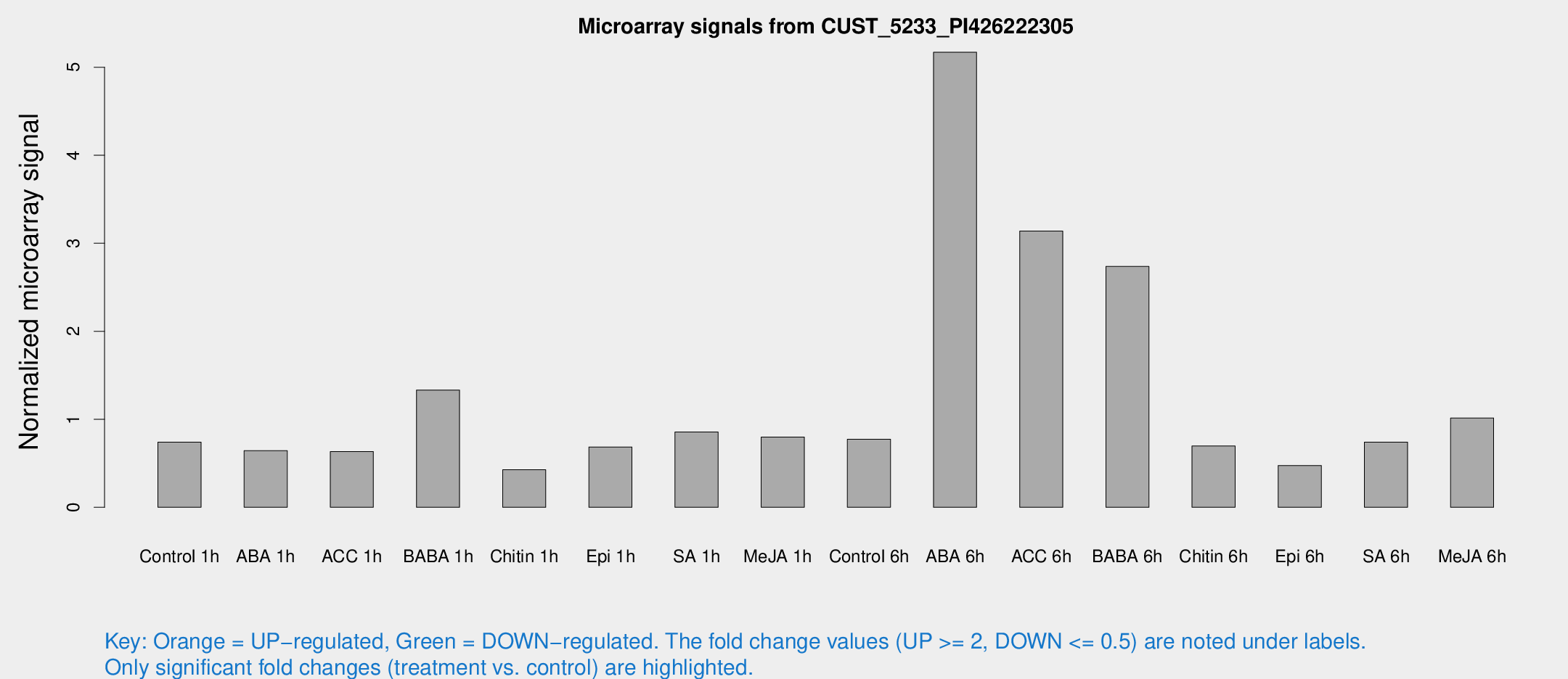
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.3961 | 3.23285 | 0.740083 | 0.108357 |
| ABA 1h | 27.0312 | 11.3174 | 0.643377 | 0.55337 |
| ACC 1h | 30.2447 | 12.3454 | 0.63371 | 0.470309 |
| BABA 1h | 43.8198 | 3.9599 | 1.33188 | 0.121175 |
| Chitin 1h | 13.8043 | 3.30241 | 0.42668 | 0.133089 |
| Epi 1h | 21.9865 | 6.49628 | 0.685529 | 0.197897 |
| SA 1h | 30.3993 | 4.43955 | 0.855129 | 0.0985821 |
| Me-JA 1h | 25.2402 | 8.61221 | 0.798897 | 0.244895 |
| Control 6h | 27.8654 | 6.74517 | 0.772416 | 0.126147 |
| ABA 6h | 204.243 | 59.4936 | 5.17035 | 1.36457 |
| ACC 6h | 123.166 | 10.907 | 3.13814 | 0.752283 |
| BABA 6h | 111.931 | 31.3465 | 2.73609 | 0.660652 |
| Chitin 6h | 27.3913 | 7.16815 | 0.696131 | 0.237293 |
| Epi 6h | 39.6135 | 31.6724 | 0.474275 | 0.794152 |
| SA 6h | 42.681 | 24.9296 | 0.740539 | 1.52195 |
| Me-JA 6h | 35.7799 | 7.68495 | 1.01437 | 0.168771 |
Source Transcript PGSC0003DMT400038581 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50660.1 | +1 | 0.0 | 580 | 288/400 (72%) | Cytochrome P450 superfamily protein | chr3:18814262-18817168 REVERSE LENGTH=513 |
