Probe CUST_5133_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5133_PI426222305 | JHI_St_60k_v1 | DMT400003882 | TATAATGGCTCTTTGGTCACCTTTAAAGCTATTGTGAGTTTCGAAAATTGCTTCTCCGAT |
All Microarray Probes Designed to Gene DMG401001531
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5133_PI426222305 | JHI_St_60k_v1 | DMT400003882 | TATAATGGCTCTTTGGTCACCTTTAAAGCTATTGTGAGTTTCGAAAATTGCTTCTCCGAT |
| CUST_5339_PI426222305 | JHI_St_60k_v1 | DMT400003883 | GGCTCTTTGGTCACCTTTAAAGCTATTAGAAAAAGCACCTGTTCCAAATAAGTCCCAAAT |
| CUST_5198_PI426222305 | JHI_St_60k_v1 | DMT400003886 | GCTCTTACTATTCAAGTTAGTGTAGTGTTAATTAAGCGTGATCAGGAGATTAATGGTTTG |
| CUST_5014_PI426222305 | JHI_St_60k_v1 | DMT400003884 | TATCTATGGTGGAGATTGAATTTGAAGAGAAAAAGCACCTGTTCCAAATAAGTCCCAAAT |
| CUST_5086_PI426222305 | JHI_St_60k_v1 | DMT400003887 | GCTCCTTGTAGGAACAAGTCTTCGAGTGATATGAGATCATGTCATGAATCATCTTTTATT |
Microarray Signals from CUST_5133_PI426222305
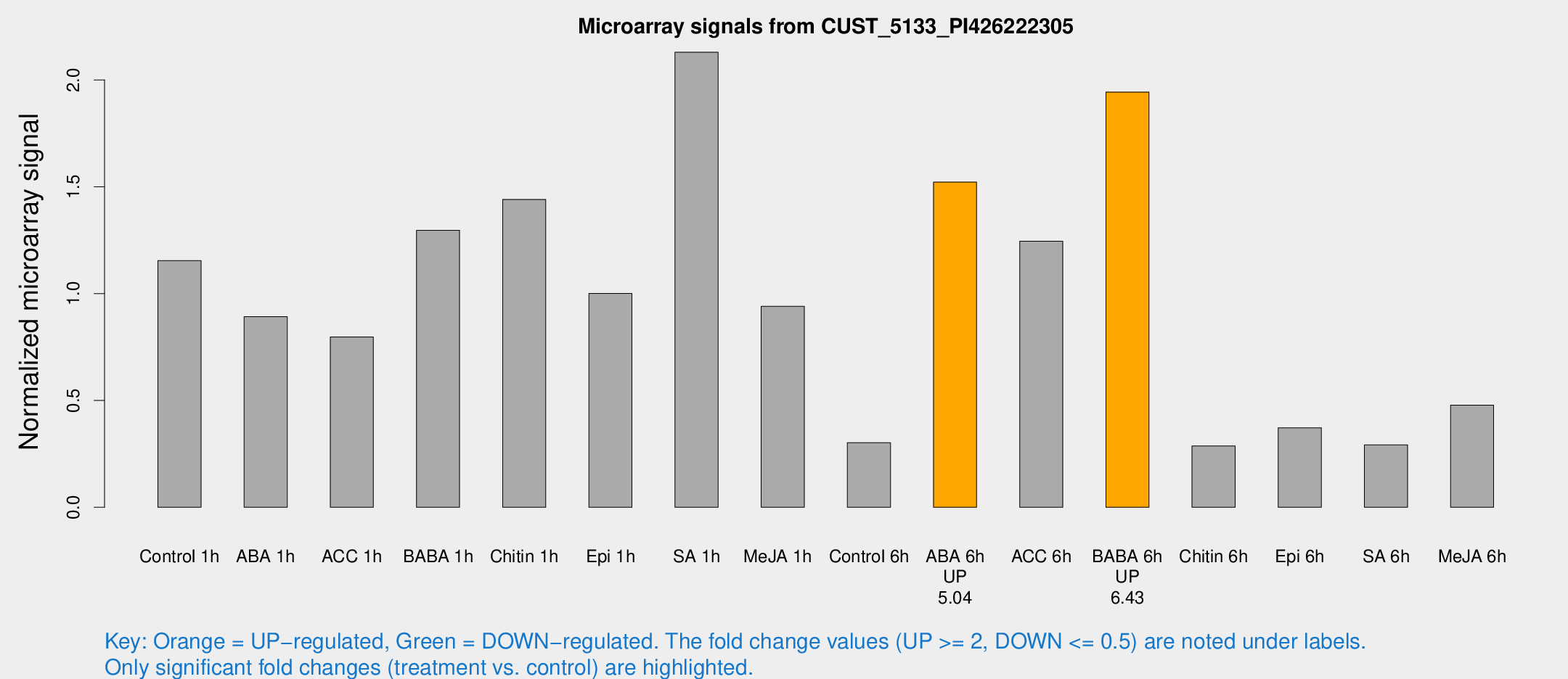
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 96.7773 | 6.59299 | 1.15414 | 0.078702 |
| ABA 1h | 67.1502 | 8.87283 | 0.892342 | 0.069152 |
| ACC 1h | 89.2884 | 35.8837 | 0.797024 | 0.485883 |
| BABA 1h | 120.794 | 37.6672 | 1.29644 | 0.431633 |
| Chitin 1h | 108.89 | 7.6534 | 1.4407 | 0.113214 |
| Epi 1h | 72.6873 | 5.40384 | 1.00094 | 0.0746437 |
| SA 1h | 186.353 | 25.4929 | 2.12968 | 0.156955 |
| Me-JA 1h | 73.8916 | 28.6879 | 0.94099 | 0.281746 |
| Control 6h | 29.6961 | 9.63616 | 0.302146 | 0.113937 |
| ABA 6h | 152.382 | 46.6952 | 1.52161 | 0.512453 |
| ACC 6h | 121.02 | 13.9634 | 1.24563 | 0.0841669 |
| BABA 6h | 196.481 | 57.2214 | 1.94333 | 0.481299 |
| Chitin 6h | 25.9028 | 4.21488 | 0.287048 | 0.0485863 |
| Epi 6h | 36.8426 | 7.4369 | 0.371718 | 0.0630161 |
| SA 6h | 35.217 | 17.9405 | 0.292017 | 0.213837 |
| Me-JA 6h | 44.2271 | 13.462 | 0.477692 | 0.125679 |
Source Transcript PGSC0003DMT400003882 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38320.1 | +2 | 6e-40 | 145 | 72/142 (51%) | TRICHOME BIREFRINGENCE-LIKE 34 | chr2:16055488-16057874 FORWARD LENGTH=410 |
