Probe CUST_5086_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5086_PI426222305 | JHI_St_60k_v1 | DMT400003887 | GCTCCTTGTAGGAACAAGTCTTCGAGTGATATGAGATCATGTCATGAATCATCTTTTATT |
All Microarray Probes Designed to Gene DMG401001531
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5133_PI426222305 | JHI_St_60k_v1 | DMT400003882 | TATAATGGCTCTTTGGTCACCTTTAAAGCTATTGTGAGTTTCGAAAATTGCTTCTCCGAT |
| CUST_5339_PI426222305 | JHI_St_60k_v1 | DMT400003883 | GGCTCTTTGGTCACCTTTAAAGCTATTAGAAAAAGCACCTGTTCCAAATAAGTCCCAAAT |
| CUST_5198_PI426222305 | JHI_St_60k_v1 | DMT400003886 | GCTCTTACTATTCAAGTTAGTGTAGTGTTAATTAAGCGTGATCAGGAGATTAATGGTTTG |
| CUST_5014_PI426222305 | JHI_St_60k_v1 | DMT400003884 | TATCTATGGTGGAGATTGAATTTGAAGAGAAAAAGCACCTGTTCCAAATAAGTCCCAAAT |
| CUST_5086_PI426222305 | JHI_St_60k_v1 | DMT400003887 | GCTCCTTGTAGGAACAAGTCTTCGAGTGATATGAGATCATGTCATGAATCATCTTTTATT |
Microarray Signals from CUST_5086_PI426222305
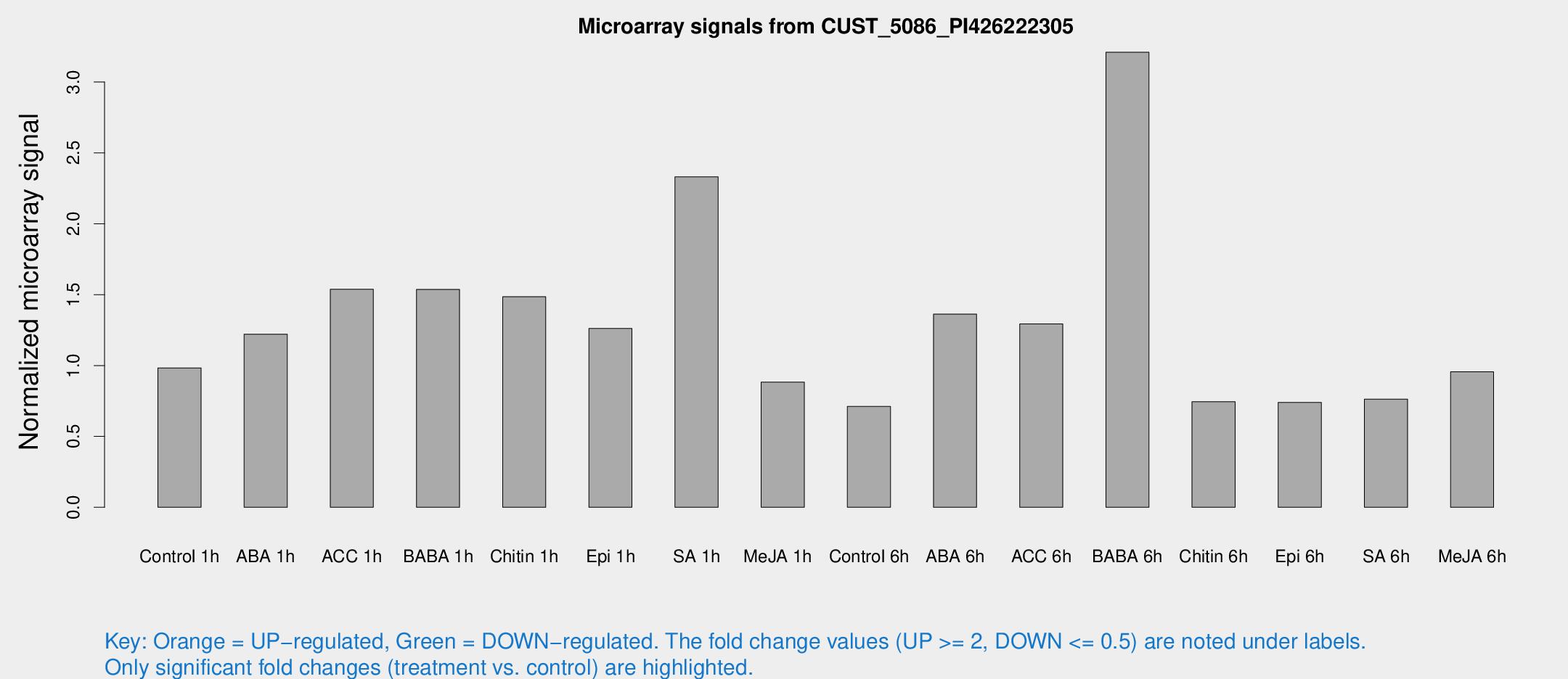
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.30224 | 3.26432 | 0.983384 | 0.442672 |
| ABA 1h | 10.3489 | 4.67231 | 1.22127 | 0.526097 |
| ACC 1h | 13.4369 | 4.02616 | 1.53822 | 0.564043 |
| BABA 1h | 14.2701 | 6.10619 | 1.53679 | 0.606929 |
| Chitin 1h | 12.8011 | 5.73573 | 1.48525 | 0.893394 |
| Epi 1h | 9.00438 | 3.20628 | 1.26109 | 0.499693 |
| SA 1h | 20.1209 | 4.79131 | 2.33064 | 0.519778 |
| Me-JA 1h | 5.74286 | 3.33576 | 0.882572 | 0.511284 |
| Control 6h | 5.66996 | 3.28577 | 0.711554 | 0.41233 |
| ABA 6h | 12.7228 | 3.74942 | 1.36268 | 0.466685 |
| ACC 6h | 12.9082 | 4.19793 | 1.29325 | 0.445954 |
| BABA 6h | 32.8034 | 12.5708 | 3.20972 | 1.14386 |
| Chitin 6h | 6.30109 | 3.65 | 0.744274 | 0.430959 |
| Epi 6h | 6.6742 | 3.89567 | 0.739506 | 0.42879 |
| SA 6h | 6.0035 | 3.48094 | 0.76287 | 0.441996 |
| Me-JA 6h | 8.29511 | 3.33893 | 0.955509 | 0.457485 |
Source Transcript PGSC0003DMT400003887 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G11030.1 | +1 | 3e-38 | 142 | 60/107 (56%) | TRICHOME BIREFRINGENCE-LIKE 32 | chr3:3457300-3459300 REVERSE LENGTH=451 |
