Probe CUST_26649_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
All Microarray Probes Designed to Gene DMG400000339
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
Microarray Signals from CUST_26649_PI426222305
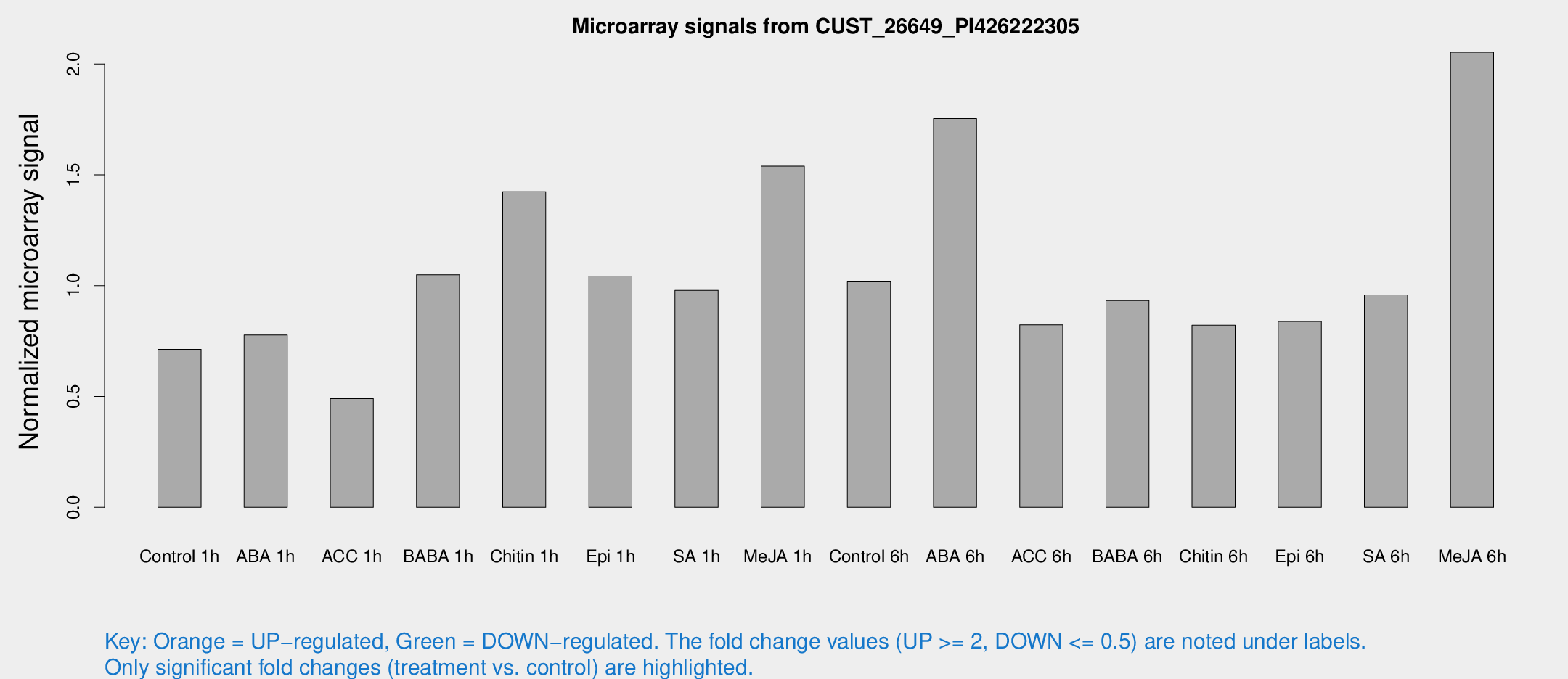
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 63.626 | 9.83846 | 0.713298 | 0.0610938 |
| ABA 1h | 60.0947 | 4.60559 | 0.777632 | 0.0689892 |
| ACC 1h | 46.8045 | 10.5737 | 0.490215 | 0.094819 |
| BABA 1h | 92.8907 | 19.3966 | 1.04941 | 0.142125 |
| Chitin 1h | 116.222 | 24.3673 | 1.42417 | 0.298111 |
| Epi 1h | 79.568 | 8.09005 | 1.0431 | 0.100499 |
| SA 1h | 87.9996 | 5.95849 | 0.979195 | 0.0661712 |
| Me-JA 1h | 113.338 | 21.3673 | 1.53896 | 0.152409 |
| Control 6h | 102.087 | 31.3753 | 1.01693 | 0.340593 |
| ABA 6h | 168.303 | 29.8609 | 1.75328 | 0.250865 |
| ACC 6h | 87.4332 | 21.7696 | 0.823609 | 0.117072 |
| BABA 6h | 98.4346 | 25.8359 | 0.93269 | 0.243283 |
| Chitin 6h | 77.4993 | 9.90421 | 0.821345 | 0.0987019 |
| Epi 6h | 86.8318 | 20.4546 | 0.838719 | 0.217252 |
| SA 6h | 88.0431 | 19.7185 | 0.958719 | 0.365449 |
| Me-JA 6h | 193.598 | 48.8509 | 2.05308 | 0.501785 |
Source Transcript PGSC0003DMT400000899 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +1 | 0.0 | 1085 | 513/705 (73%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
