Probe CUST_26587_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
All Microarray Probes Designed to Gene DMG400000339
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
Microarray Signals from CUST_26587_PI426222305
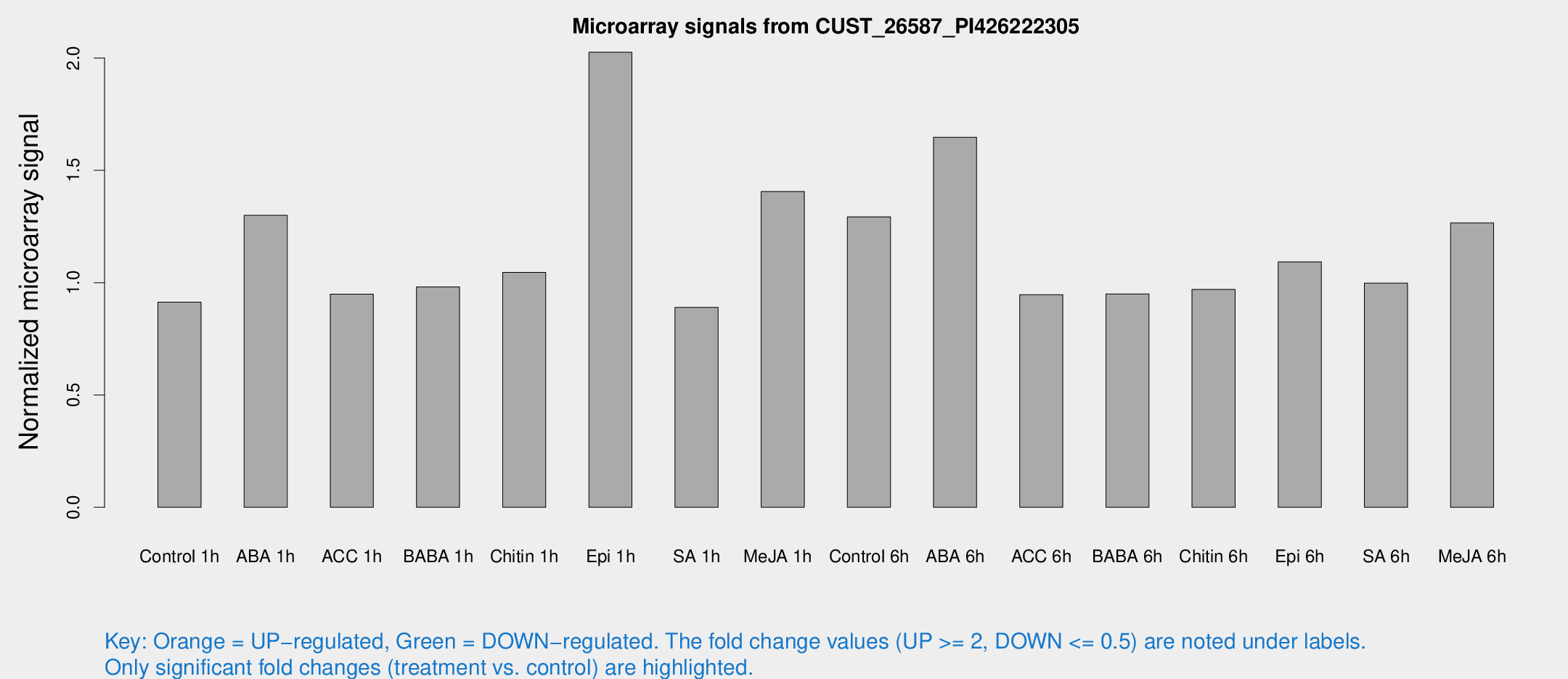
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4.88935 | 2.83542 | 0.912908 | 0.515815 |
| ABA 1h | 6.58128 | 2.76644 | 1.29954 | 0.609882 |
| ACC 1h | 5.35889 | 3.10762 | 0.948706 | 0.549371 |
| BABA 1h | 5.18453 | 3.01104 | 0.980856 | 0.567297 |
| Chitin 1h | 4.92474 | 2.86964 | 1.04556 | 0.576819 |
| Epi 1h | 10.4794 | 2.91068 | 2.0256 | 0.721563 |
| SA 1h | 4.9438 | 2.86603 | 0.88985 | 0.507822 |
| Me-JA 1h | 6.41386 | 2.94229 | 1.40551 | 0.680324 |
| Control 6h | 7.99064 | 3.00282 | 1.29228 | 0.60914 |
| ABA 6h | 10.4194 | 3.14995 | 1.64728 | 0.609996 |
| ACC 6h | 6.08712 | 3.6017 | 0.946451 | 0.548062 |
| BABA 6h | 5.86138 | 3.35664 | 0.949645 | 0.543355 |
| Chitin 6h | 5.67042 | 3.29179 | 0.969593 | 0.562743 |
| Epi 6h | 6.93078 | 3.42922 | 1.09209 | 0.558668 |
| SA 6h | 5.42396 | 3.1455 | 0.997667 | 0.578169 |
| Me-JA 6h | 7.05983 | 3.00208 | 1.26576 | 0.560519 |
Source Transcript PGSC0003DMT400000896 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +2 | 1e-13 | 72 | 33/50 (66%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
