Probe CUST_26564_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
All Microarray Probes Designed to Gene DMG400000339
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
Microarray Signals from CUST_26564_PI426222305
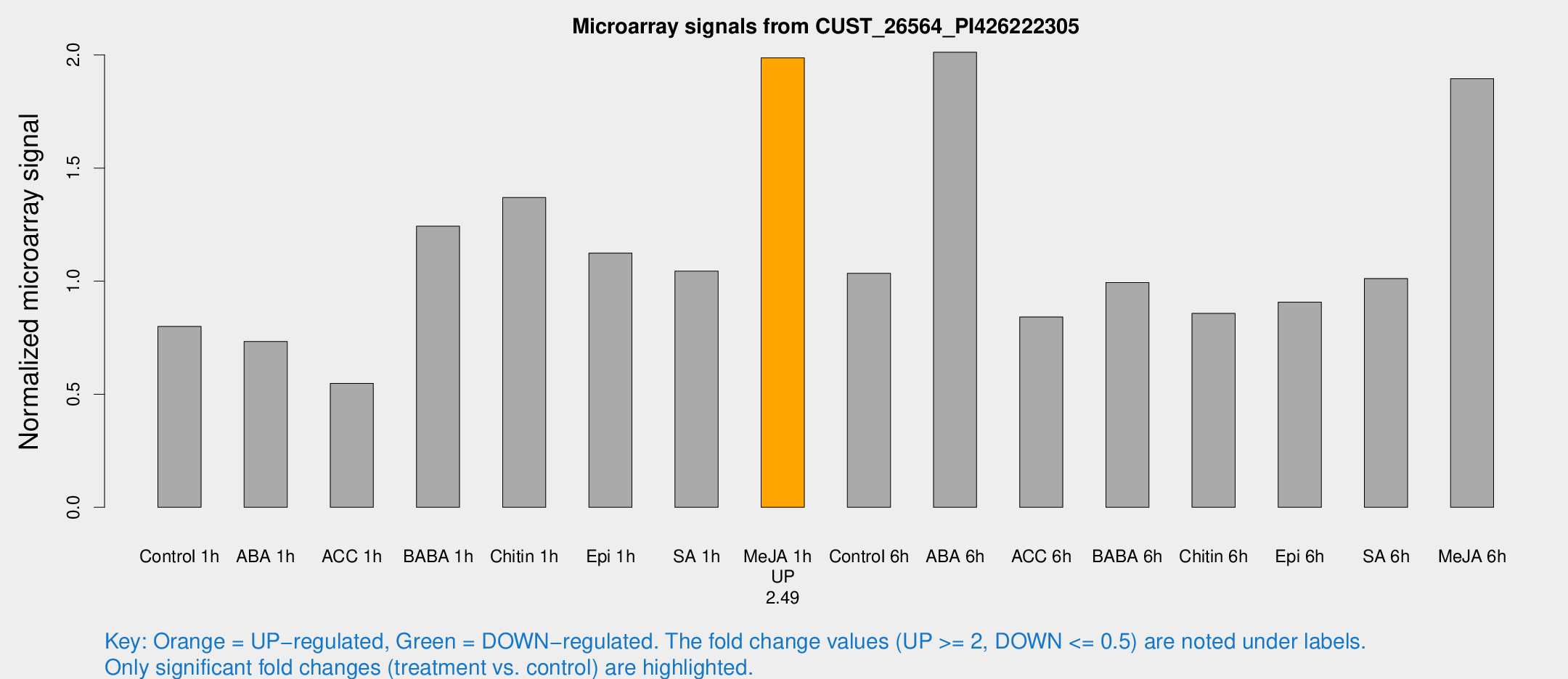
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 146.141 | 17.2097 | 0.799471 | 0.0646039 |
| ABA 1h | 120.291 | 21.8503 | 0.732752 | 0.137746 |
| ACC 1h | 108.145 | 25.7949 | 0.547666 | 0.105589 |
| BABA 1h | 225.984 | 45.6672 | 1.24272 | 0.154149 |
| Chitin 1h | 223.902 | 22.1557 | 1.36984 | 0.135092 |
| Epi 1h | 177.278 | 19.0732 | 1.12385 | 0.104122 |
| SA 1h | 195.011 | 19.0944 | 1.04385 | 0.141943 |
| Me-JA 1h | 294.344 | 26.1683 | 1.98694 | 0.116676 |
| Control 6h | 211.525 | 62.7262 | 1.03432 | 0.316344 |
| ABA 6h | 396.077 | 68.3955 | 2.01195 | 0.226062 |
| ACC 6h | 175.36 | 16.7904 | 0.84174 | 0.0517337 |
| BABA 6h | 208.712 | 42.6567 | 0.993363 | 0.167575 |
| Chitin 6h | 166.795 | 19.4877 | 0.85737 | 0.0860485 |
| Epi 6h | 194.032 | 46.2165 | 0.907486 | 0.203157 |
| SA 6h | 181.404 | 14.725 | 1.01161 | 0.178521 |
| Me-JA 6h | 356.302 | 70.5223 | 1.89529 | 0.299586 |
Source Transcript PGSC0003DMT400000898 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +2 | 0.0 | 897 | 428/600 (71%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
