Probe CUST_26602_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
All Microarray Probes Designed to Gene DMG400000339
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
Microarray Signals from CUST_26602_PI426222305
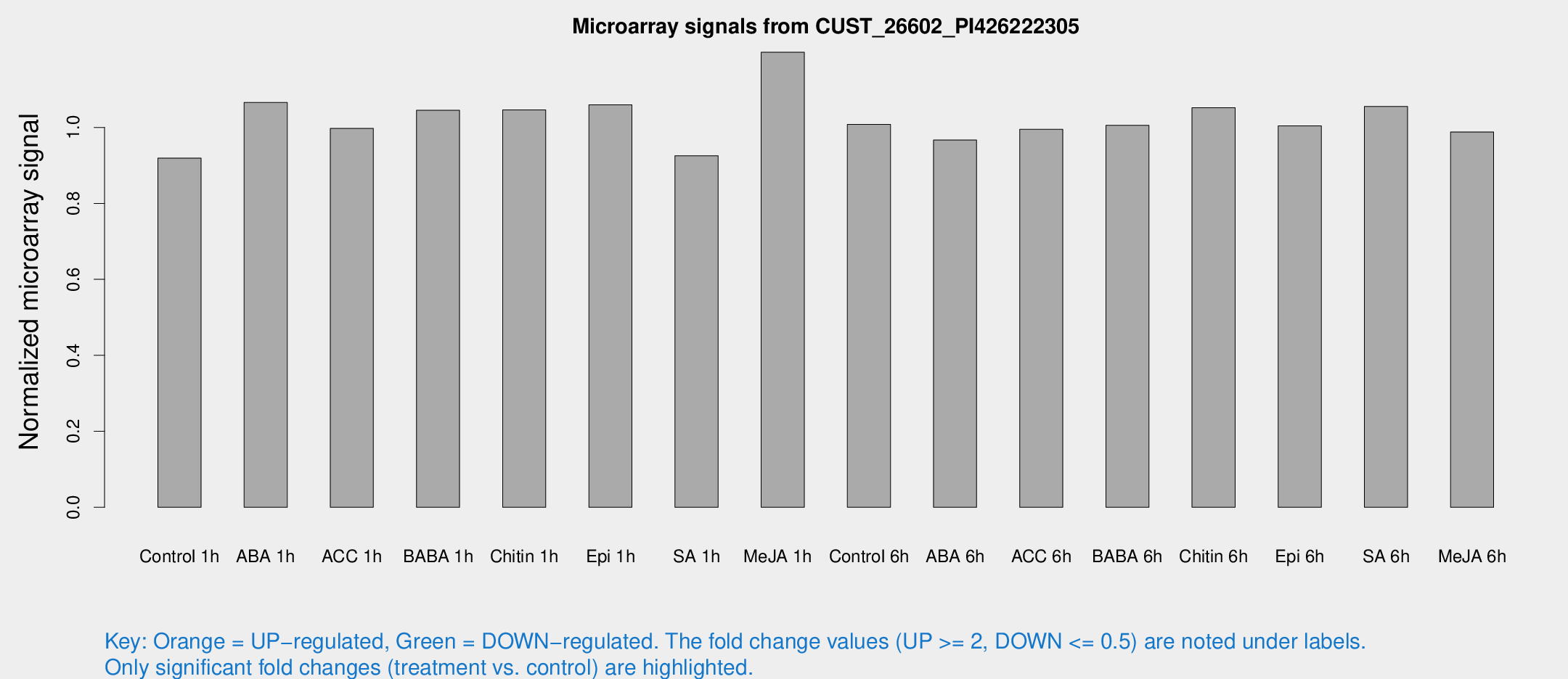
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.05676 | 3.50924 | 0.919274 | 0.532316 |
| ABA 1h | 6.22186 | 3.48955 | 1.06563 | 0.598563 |
| ACC 1h | 6.76043 | 3.92433 | 0.99728 | 0.577558 |
| BABA 1h | 6.63688 | 3.84512 | 1.04516 | 0.605249 |
| Chitin 1h | 6.21617 | 3.61279 | 1.04618 | 0.605986 |
| Epi 1h | 6.04727 | 3.51071 | 1.05931 | 0.614035 |
| SA 1h | 6.26205 | 3.63269 | 0.925388 | 0.536688 |
| Me-JA 1h | 6.46435 | 3.75834 | 1.19794 | 0.693794 |
| Control 6h | 6.65399 | 3.81029 | 1.008 | 0.577374 |
| ABA 6h | 6.78048 | 3.93707 | 0.966648 | 0.560175 |
| ACC 6h | 7.66954 | 4.53731 | 0.994839 | 0.576082 |
| BABA 6h | 7.44111 | 4.32951 | 1.00548 | 0.582862 |
| Chitin 6h | 7.37499 | 4.27087 | 1.05197 | 0.609115 |
| Epi 6h | 7.48883 | 4.35878 | 1.00393 | 0.581661 |
| SA 6h | 6.87724 | 3.98544 | 1.05505 | 0.610959 |
| Me-JA 6h | 6.4855 | 3.75888 | 0.988124 | 0.572189 |
Source Transcript PGSC0003DMT400000897 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +2 | 2e-168 | 504 | 252/365 (69%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
