Probe CUST_26570_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
All Microarray Probes Designed to Gene DMG400000339
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26587_PI426222305 | JHI_St_60k_v1 | DMT400000896 | CCTTTGACAACACTGATCTTGATTCAAAATTATGTATTTCGCGTCACGTCTAAATCCATA |
| CUST_26649_PI426222305 | JHI_St_60k_v1 | DMT400000899 | CATCAAATTGGCACAAAGAGCAGGACTTTATGTCAATTTACGTATTGGCCCTTACATTTG |
| CUST_26570_PI426222305 | JHI_St_60k_v1 | DMT400000900 | TGGAGGCTTAGATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGG |
| CUST_26602_PI426222305 | JHI_St_60k_v1 | DMT400000897 | CAGATCCACTATGTGAGACCTACACTCGATATGTTTTTCTTCTAACTCTGTTTACATTTT |
| CUST_26564_PI426222305 | JHI_St_60k_v1 | DMT400000898 | GATGTTATTGAAACTTATGTTTTCTGGAATGCACATGAGCCTTCTCCTGGAAAAAAGAAA |
Microarray Signals from CUST_26570_PI426222305
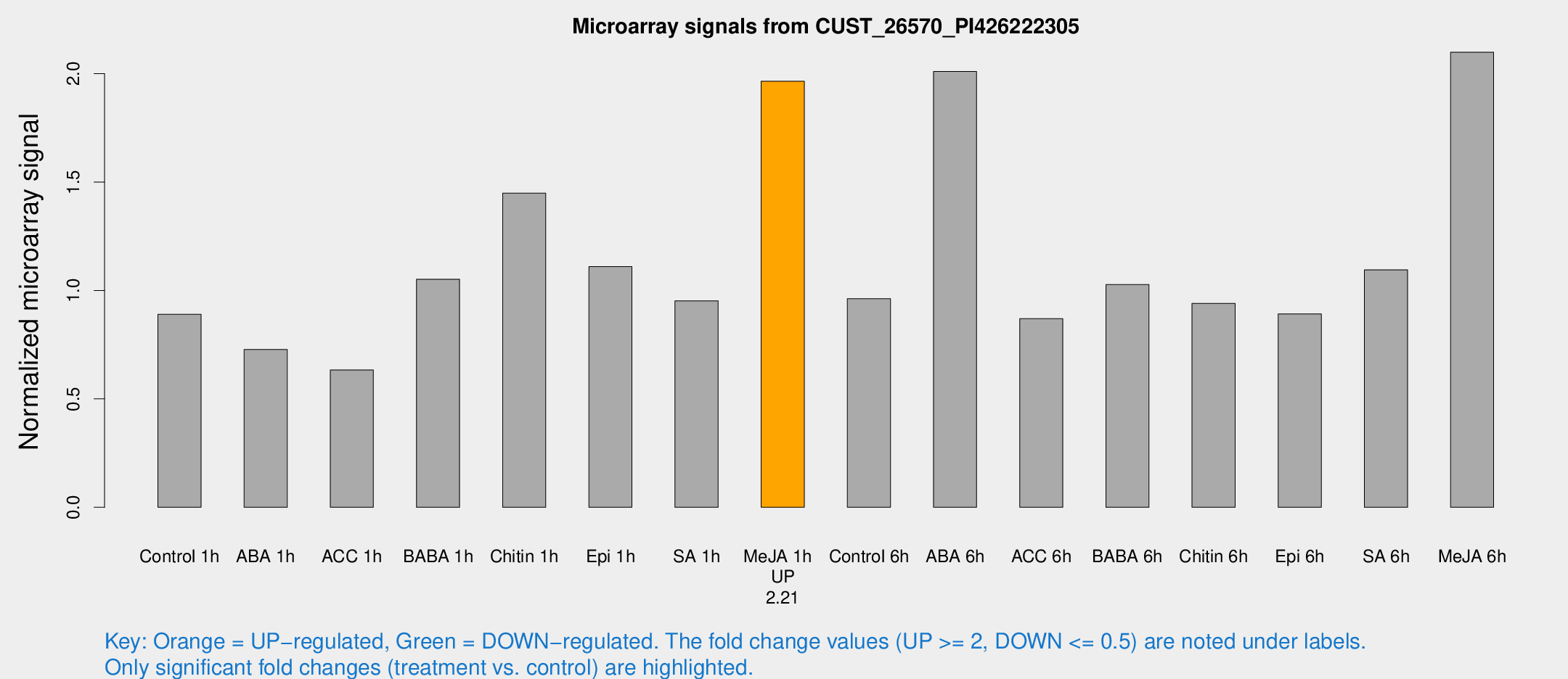
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 141.881 | 13.8892 | 0.890051 | 0.0564366 |
| ABA 1h | 103.679 | 14.6354 | 0.727849 | 0.0931107 |
| ACC 1h | 104.812 | 14.3986 | 0.633266 | 0.0448574 |
| BABA 1h | 173.758 | 46.5634 | 1.05165 | 0.216767 |
| Chitin 1h | 215.97 | 50.403 | 1.44842 | 0.34516 |
| Epi 1h | 153.706 | 18.0125 | 1.1097 | 0.106327 |
| SA 1h | 155.848 | 15.2027 | 0.952093 | 0.135197 |
| Me-JA 1h | 253.961 | 15.1834 | 1.9645 | 0.117014 |
| Control 6h | 178.99 | 58.2694 | 0.961684 | 0.368004 |
| ABA 6h | 350.018 | 68.5979 | 2.00958 | 0.278158 |
| ACC 6h | 158.359 | 11.6154 | 0.869778 | 0.0857763 |
| BABA 6h | 186.227 | 30.2612 | 1.02715 | 0.127072 |
| Chitin 6h | 158.225 | 10.189 | 0.940556 | 0.0605303 |
| Epi 6h | 171.107 | 46.825 | 0.891994 | 0.295874 |
| SA 6h | 171.702 | 11.7265 | 1.0949 | 0.208164 |
| Me-JA 6h | 348.997 | 77.756 | 2.0986 | 0.408433 |
Source Transcript PGSC0003DMT400000900 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +2 | 0.0 | 832 | 398/567 (70%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
