Probe CUST_49923_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49923_PI426222305 | JHI_St_60k_v1 | DMT400062205 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
All Microarray Probes Designed to Gene DMG400024209
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49922_PI426222305 | JHI_St_60k_v1 | DMT400062203 | AGAGTTGACACAGCTATATTTTGATAACCCTGACTAAAGAATTTGAACTTGTATCAGTCG |
| CUST_49919_PI426222305 | JHI_St_60k_v1 | DMT400062204 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49928_PI426222305 | JHI_St_60k_v1 | DMT400062210 | GACGCCTTGTATCACCGGCCTTTCTTATTTACTGATTAATTTTGTTAAATGTACTTGGTA |
| CUST_49923_PI426222305 | JHI_St_60k_v1 | DMT400062205 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49920_PI426222305 | JHI_St_60k_v1 | DMT400062211 | AGGAAGCTTTGTACAATGCTGGCAGATTGACAGGTTAGTCCTTAATACGACAACACATTT |
Microarray Signals from CUST_49923_PI426222305
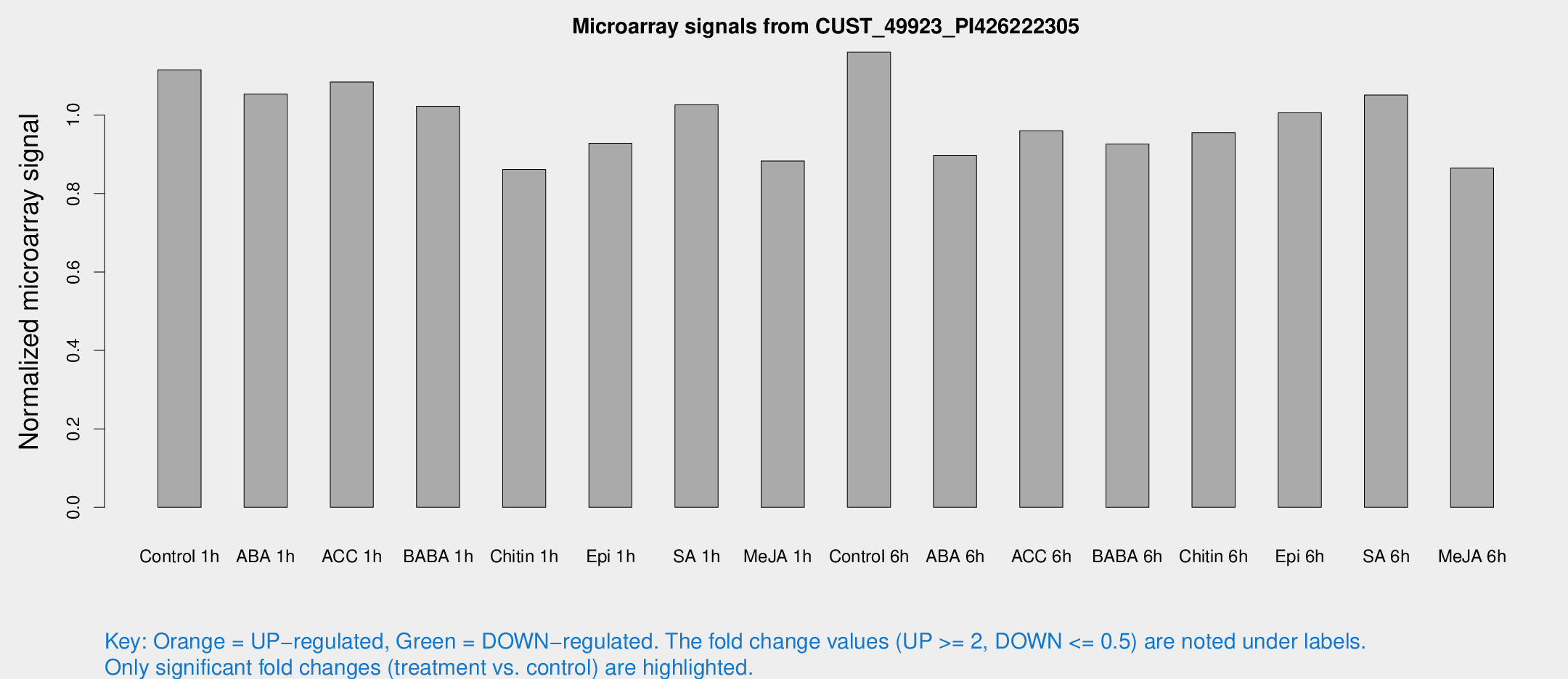
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 237.306 | 14.1698 | 1.11565 | 0.0664989 |
| ABA 1h | 198.123 | 11.9466 | 1.05338 | 0.0640686 |
| ACC 1h | 239.283 | 25.4638 | 1.08448 | 0.0665803 |
| BABA 1h | 214.103 | 33.3888 | 1.0224 | 0.0989311 |
| Chitin 1h | 165.06 | 10.2316 | 0.86169 | 0.0660556 |
| Epi 1h | 170.763 | 10.4429 | 0.928267 | 0.0567632 |
| SA 1h | 225.694 | 21.7939 | 1.02618 | 0.113554 |
| Me-JA 1h | 155.162 | 17.7479 | 0.882997 | 0.0708455 |
| Control 6h | 262.944 | 62.7036 | 1.1604 | 0.215535 |
| ABA 6h | 206.002 | 27.9361 | 0.896849 | 0.0698969 |
| ACC 6h | 234.765 | 15.4311 | 0.960178 | 0.0761984 |
| BABA 6h | 222.97 | 25.6281 | 0.926568 | 0.0899449 |
| Chitin 6h | 216.86 | 14.845 | 0.955495 | 0.0587191 |
| Epi 6h | 245.609 | 35.5929 | 1.00616 | 0.134689 |
| SA 6h | 227.201 | 36.8889 | 1.05115 | 0.302845 |
| Me-JA 6h | 189.151 | 33.4477 | 0.865127 | 0.0897546 |
Source Transcript PGSC0003DMT400062205 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G50330.1 | +1 | 0.0 | 599 | 304/453 (67%) | Protein kinase superfamily protein | chr5:20485406-20488563 REVERSE LENGTH=479 |
