Probe CUST_49919_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49919_PI426222305 | JHI_St_60k_v1 | DMT400062204 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
All Microarray Probes Designed to Gene DMG400024209
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49922_PI426222305 | JHI_St_60k_v1 | DMT400062203 | AGAGTTGACACAGCTATATTTTGATAACCCTGACTAAAGAATTTGAACTTGTATCAGTCG |
| CUST_49919_PI426222305 | JHI_St_60k_v1 | DMT400062204 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49928_PI426222305 | JHI_St_60k_v1 | DMT400062210 | GACGCCTTGTATCACCGGCCTTTCTTATTTACTGATTAATTTTGTTAAATGTACTTGGTA |
| CUST_49923_PI426222305 | JHI_St_60k_v1 | DMT400062205 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49920_PI426222305 | JHI_St_60k_v1 | DMT400062211 | AGGAAGCTTTGTACAATGCTGGCAGATTGACAGGTTAGTCCTTAATACGACAACACATTT |
Microarray Signals from CUST_49919_PI426222305
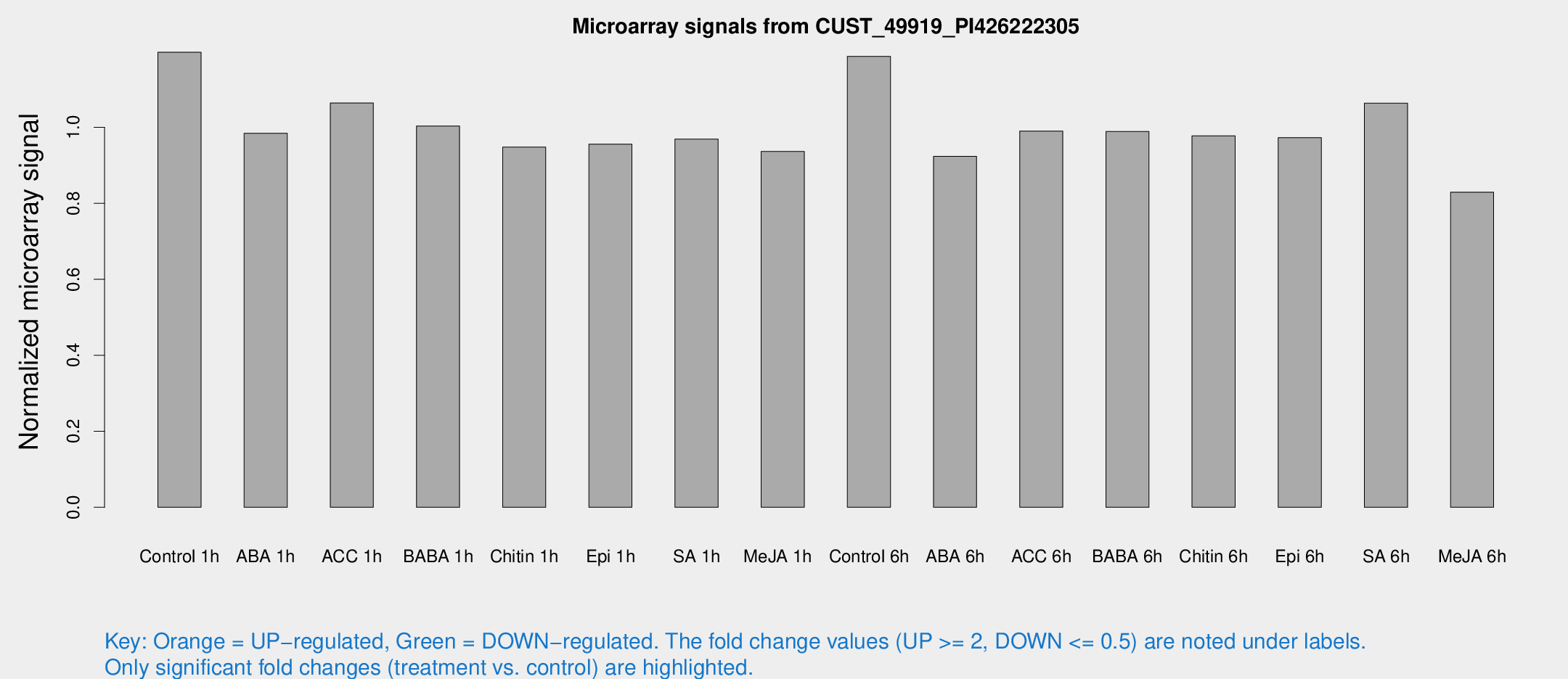
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 251.153 | 15.2058 | 1.19744 | 0.0708885 |
| ABA 1h | 182.214 | 11.017 | 0.984294 | 0.0594277 |
| ACC 1h | 232.088 | 28.403 | 1.06371 | 0.0639246 |
| BABA 1h | 209.933 | 41.8585 | 1.00336 | 0.132632 |
| Chitin 1h | 180.585 | 20.7355 | 0.947848 | 0.0869145 |
| Epi 1h | 173.914 | 13.6086 | 0.955618 | 0.073921 |
| SA 1h | 208.851 | 13.2869 | 0.968812 | 0.122333 |
| Me-JA 1h | 159.807 | 9.85054 | 0.936303 | 0.0712813 |
| Control 6h | 266.057 | 67.9978 | 1.18668 | 0.239811 |
| ABA 6h | 206.098 | 13.6755 | 0.923407 | 0.0558059 |
| ACC 6h | 237.717 | 14.3969 | 0.989971 | 0.0974911 |
| BABA 6h | 234.4 | 27.0101 | 0.988938 | 0.0944673 |
| Chitin 6h | 217.779 | 13.1743 | 0.977532 | 0.0660461 |
| Epi 6h | 236.393 | 42.7056 | 0.972912 | 0.1988 |
| SA 6h | 224.585 | 31.6648 | 1.06335 | 0.277986 |
| Me-JA 6h | 181.635 | 37.2094 | 0.829214 | 0.134872 |
Source Transcript PGSC0003DMT400062204 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24810.2 | +2 | 4e-40 | 146 | 67/100 (67%) | Protein kinase superfamily protein | chr4:12786791-12789598 REVERSE LENGTH=481 |
