Probe CUST_49920_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49920_PI426222305 | JHI_St_60k_v1 | DMT400062211 | AGGAAGCTTTGTACAATGCTGGCAGATTGACAGGTTAGTCCTTAATACGACAACACATTT |
All Microarray Probes Designed to Gene DMG400024209
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49922_PI426222305 | JHI_St_60k_v1 | DMT400062203 | AGAGTTGACACAGCTATATTTTGATAACCCTGACTAAAGAATTTGAACTTGTATCAGTCG |
| CUST_49919_PI426222305 | JHI_St_60k_v1 | DMT400062204 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49928_PI426222305 | JHI_St_60k_v1 | DMT400062210 | GACGCCTTGTATCACCGGCCTTTCTTATTTACTGATTAATTTTGTTAAATGTACTTGGTA |
| CUST_49923_PI426222305 | JHI_St_60k_v1 | DMT400062205 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49920_PI426222305 | JHI_St_60k_v1 | DMT400062211 | AGGAAGCTTTGTACAATGCTGGCAGATTGACAGGTTAGTCCTTAATACGACAACACATTT |
Microarray Signals from CUST_49920_PI426222305
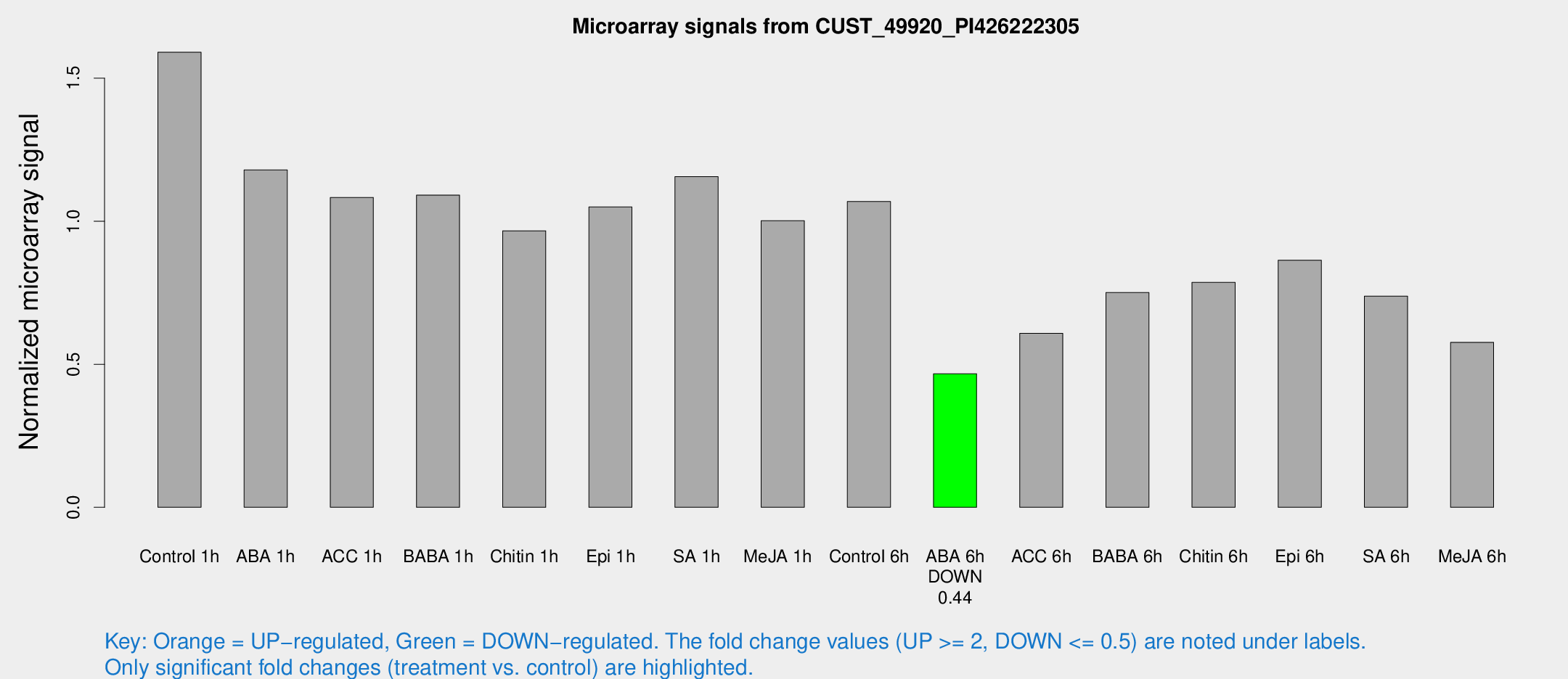
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1230.93 | 163.641 | 1.5906 | 0.14504 |
| ABA 1h | 797.378 | 52.9595 | 1.17918 | 0.133004 |
| ACC 1h | 894.544 | 189.483 | 1.08281 | 0.185582 |
| BABA 1h | 814.747 | 104.7 | 1.09118 | 0.0632321 |
| Chitin 1h | 665.514 | 53.3941 | 0.96655 | 0.119078 |
| Epi 1h | 692.183 | 40.1731 | 1.04967 | 0.0608553 |
| SA 1h | 904.809 | 52.4469 | 1.15568 | 0.104685 |
| Me-JA 1h | 627.44 | 57.598 | 1.00171 | 0.0581543 |
| Control 6h | 872.741 | 215.488 | 1.06884 | 0.209837 |
| ABA 6h | 378.53 | 22.2816 | 0.466676 | 0.027409 |
| ACC 6h | 543.753 | 83.8538 | 0.607882 | 0.0380442 |
| BABA 6h | 659.58 | 118.516 | 0.750617 | 0.120013 |
| Chitin 6h | 638.155 | 37.1782 | 0.786173 | 0.0457077 |
| Epi 6h | 752.569 | 93.2629 | 0.863809 | 0.145928 |
| SA 6h | 591.798 | 154.35 | 0.73812 | 0.138277 |
| Me-JA 6h | 453.194 | 85.4931 | 0.57642 | 0.0629254 |
Source Transcript PGSC0003DMT400062211 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24810.2 | +2 | 1e-41 | 146 | 67/100 (67%) | Protein kinase superfamily protein | chr4:12786791-12789598 REVERSE LENGTH=481 |
