Probe CUST_49922_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49922_PI426222305 | JHI_St_60k_v1 | DMT400062203 | AGAGTTGACACAGCTATATTTTGATAACCCTGACTAAAGAATTTGAACTTGTATCAGTCG |
All Microarray Probes Designed to Gene DMG400024209
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49922_PI426222305 | JHI_St_60k_v1 | DMT400062203 | AGAGTTGACACAGCTATATTTTGATAACCCTGACTAAAGAATTTGAACTTGTATCAGTCG |
| CUST_49919_PI426222305 | JHI_St_60k_v1 | DMT400062204 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49928_PI426222305 | JHI_St_60k_v1 | DMT400062210 | GACGCCTTGTATCACCGGCCTTTCTTATTTACTGATTAATTTTGTTAAATGTACTTGGTA |
| CUST_49923_PI426222305 | JHI_St_60k_v1 | DMT400062205 | CATACACAACCCCTCAAACTTGTCCTCATTTTTCATTTTGGCACTCTAATTTAGCCTTGT |
| CUST_49920_PI426222305 | JHI_St_60k_v1 | DMT400062211 | AGGAAGCTTTGTACAATGCTGGCAGATTGACAGGTTAGTCCTTAATACGACAACACATTT |
Microarray Signals from CUST_49922_PI426222305
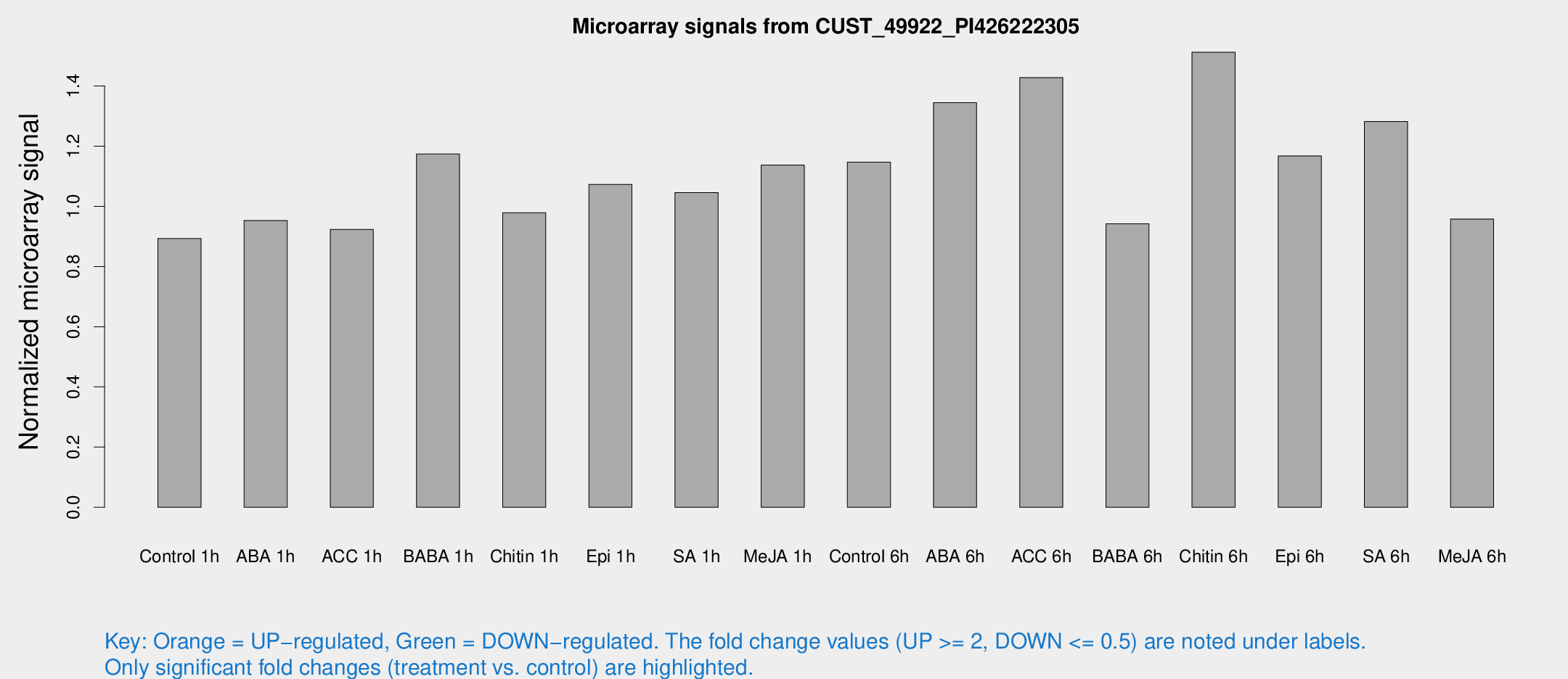
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.08627 | 3.53053 | 0.893036 | 0.517996 |
| ABA 1h | 5.74698 | 3.33163 | 0.952694 | 0.551935 |
| ACC 1h | 6.47079 | 3.75377 | 0.923503 | 0.535114 |
| BABA 1h | 8.08816 | 3.77958 | 1.17371 | 0.592336 |
| Chitin 1h | 6.02412 | 3.50595 | 0.978653 | 0.567011 |
| Epi 1h | 6.40038 | 3.44208 | 1.07302 | 0.58387 |
| SA 1h | 7.80099 | 3.40286 | 1.0457 | 0.507531 |
| Me-JA 1h | 6.38671 | 3.60054 | 1.13708 | 0.641233 |
| Control 6h | 8.2484 | 3.70009 | 1.14698 | 0.567239 |
| ABA 6h | 11.0857 | 4.1826 | 1.34426 | 0.632589 |
| ACC 6h | 12.1302 | 4.3714 | 1.42738 | 0.59878 |
| BABA 6h | 7.2588 | 4.25352 | 0.942062 | 0.546103 |
| Chitin 6h | 12.2809 | 4.49811 | 1.51202 | 0.706602 |
| Epi 6h | 9.23232 | 4.354 | 1.16747 | 0.579431 |
| SA 6h | 9.14571 | 4.08438 | 1.28168 | 0.633985 |
| Me-JA 6h | 6.50967 | 3.7805 | 0.957707 | 0.555241 |
Source Transcript PGSC0003DMT400062203 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24810.2 | +2 | 1e-16 | 77 | 34/52 (65%) | Protein kinase superfamily protein | chr4:12786791-12789598 REVERSE LENGTH=481 |
