Probe CUST_46253_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46253_PI426222305 | JHI_St_60k_v1 | DMT400053236 | AGGGTTTAATTTCTAACTGCATTGTAACTGTGCAAGTGTTCTAATGAATGAAATCCCTTC |
All Microarray Probes Designed to Gene DMG401020664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46274_PI426222305 | JHI_St_60k_v1 | DMT400053235 | ATTCTGGTACTGCATCATCGACATTTTGCATTCCGACACCATTTATCATTTACTAACATT |
| CUST_46236_PI426222305 | JHI_St_60k_v1 | DMT400053238 | GATTTCGGAGGTGACATGAGAGAAAGTGAATTATATTGAGGGACGTAAGTTACTATTGAA |
| CUST_46240_PI426222305 | JHI_St_60k_v1 | DMT400053237 | CTAACATCTTGTTCATTTCTCTGTTTCTCAGGAGGTGACCTGAGAGAAAGTGAGTTATAT |
| CUST_46253_PI426222305 | JHI_St_60k_v1 | DMT400053236 | AGGGTTTAATTTCTAACTGCATTGTAACTGTGCAAGTGTTCTAATGAATGAAATCCCTTC |
| CUST_46246_PI426222305 | JHI_St_60k_v1 | DMT400053239 | CAAGCAGAACAAGCTGTTATCGATGCGCTTCCTTAAAAAGTGATTATTATGGTATTGGGG |
Microarray Signals from CUST_46253_PI426222305
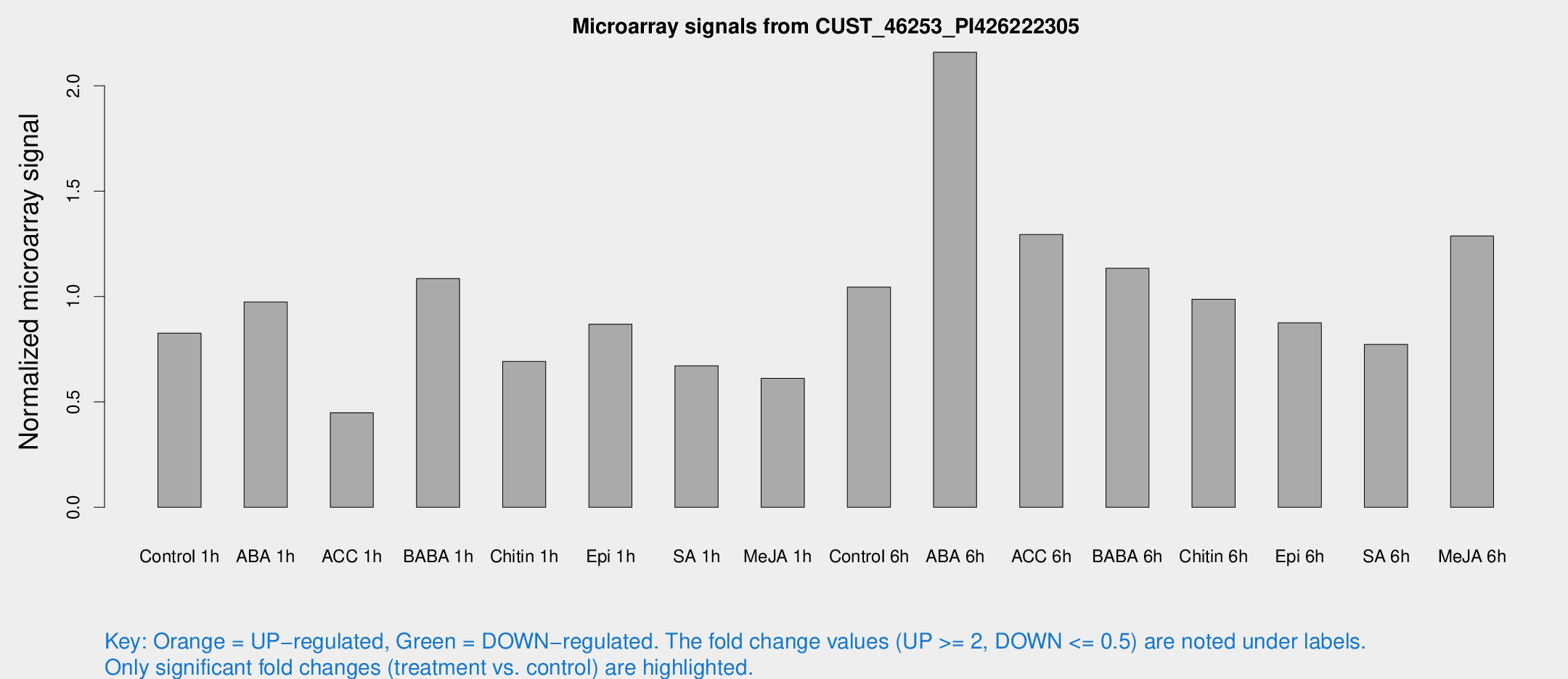
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 256.322 | 44.9672 | 0.826628 | 0.209589 |
| ABA 1h | 315.636 | 134.209 | 0.974762 | 0.427038 |
| ACC 1h | 237.145 | 109.179 | 0.448937 | 0.682042 |
| BABA 1h | 332.068 | 75.5182 | 1.0855 | 0.165878 |
| Chitin 1h | 200.173 | 55.72 | 0.692521 | 0.141243 |
| Epi 1h | 228.939 | 29.629 | 0.868901 | 0.114834 |
| SA 1h | 239.132 | 77.378 | 0.67168 | 0.272474 |
| Me-JA 1h | 153.757 | 23.5516 | 0.612287 | 0.127439 |
| Control 6h | 321.369 | 49.5686 | 1.04523 | 0.172634 |
| ABA 6h | 697.692 | 79.609 | 2.15924 | 0.299121 |
| ACC 6h | 451.877 | 56.5226 | 1.29412 | 0.112473 |
| BABA 6h | 392.101 | 63.6394 | 1.13426 | 0.165659 |
| Chitin 6h | 326.573 | 64.1299 | 0.986941 | 0.206217 |
| Epi 6h | 301.467 | 39.6481 | 0.875363 | 0.192417 |
| SA 6h | 253.955 | 69.9001 | 0.772688 | 0.180547 |
| Me-JA 6h | 420.464 | 130.626 | 1.28781 | 0.288996 |
Source Transcript PGSC0003DMT400053236 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26695.1 | +2 | 1e-63 | 197 | 87/136 (64%) | Ran BP2/NZF zinc finger-like superfamily protein | chr2:11365275-11365789 FORWARD LENGTH=138 |
