Probe CUST_46236_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46236_PI426222305 | JHI_St_60k_v1 | DMT400053238 | GATTTCGGAGGTGACATGAGAGAAAGTGAATTATATTGAGGGACGTAAGTTACTATTGAA |
All Microarray Probes Designed to Gene DMG401020664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46274_PI426222305 | JHI_St_60k_v1 | DMT400053235 | ATTCTGGTACTGCATCATCGACATTTTGCATTCCGACACCATTTATCATTTACTAACATT |
| CUST_46236_PI426222305 | JHI_St_60k_v1 | DMT400053238 | GATTTCGGAGGTGACATGAGAGAAAGTGAATTATATTGAGGGACGTAAGTTACTATTGAA |
| CUST_46240_PI426222305 | JHI_St_60k_v1 | DMT400053237 | CTAACATCTTGTTCATTTCTCTGTTTCTCAGGAGGTGACCTGAGAGAAAGTGAGTTATAT |
| CUST_46253_PI426222305 | JHI_St_60k_v1 | DMT400053236 | AGGGTTTAATTTCTAACTGCATTGTAACTGTGCAAGTGTTCTAATGAATGAAATCCCTTC |
| CUST_46246_PI426222305 | JHI_St_60k_v1 | DMT400053239 | CAAGCAGAACAAGCTGTTATCGATGCGCTTCCTTAAAAAGTGATTATTATGGTATTGGGG |
Microarray Signals from CUST_46236_PI426222305
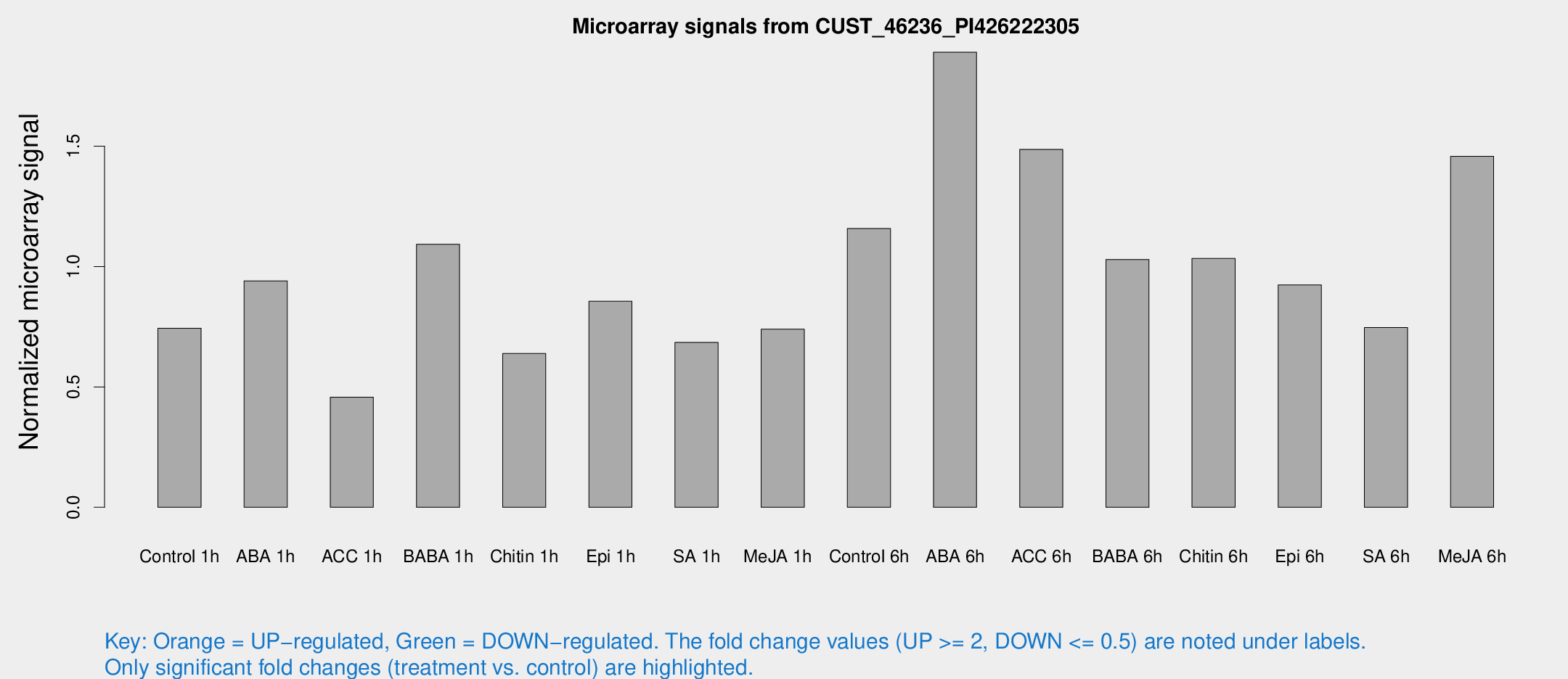
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 286.581 | 51.6052 | 0.74387 | 0.188058 |
| ABA 1h | 381.666 | 177.445 | 0.940494 | 0.401924 |
| ACC 1h | 275.53 | 124.597 | 0.457529 | 0.527777 |
| BABA 1h | 405.494 | 74.3018 | 1.09267 | 0.114606 |
| Chitin 1h | 235.962 | 77.9111 | 0.638723 | 0.155424 |
| Epi 1h | 279.72 | 35.7695 | 0.855858 | 0.116031 |
| SA 1h | 314.805 | 115.591 | 0.684939 | 0.331689 |
| Me-JA 1h | 227.537 | 25.8608 | 0.740036 | 0.135104 |
| Control 6h | 440.396 | 62.7849 | 1.15789 | 0.183369 |
| ABA 6h | 782.282 | 161.913 | 1.89027 | 0.480612 |
| ACC 6h | 648.794 | 91.6929 | 1.48686 | 0.244722 |
| BABA 6h | 436.437 | 56.6846 | 1.0296 | 0.11008 |
| Chitin 6h | 439.245 | 116.564 | 1.03341 | 0.312501 |
| Epi 6h | 416.594 | 111.219 | 0.923676 | 0.353266 |
| SA 6h | 316.931 | 107.307 | 0.746961 | 0.227618 |
| Me-JA 6h | 640.711 | 277.942 | 1.45802 | 0.525512 |
Source Transcript PGSC0003DMT400053238 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26695.1 | +2 | 2e-63 | 196 | 87/136 (64%) | Ran BP2/NZF zinc finger-like superfamily protein | chr2:11365275-11365789 FORWARD LENGTH=138 |
