Probe CUST_46246_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46246_PI426222305 | JHI_St_60k_v1 | DMT400053239 | CAAGCAGAACAAGCTGTTATCGATGCGCTTCCTTAAAAAGTGATTATTATGGTATTGGGG |
All Microarray Probes Designed to Gene DMG401020664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46274_PI426222305 | JHI_St_60k_v1 | DMT400053235 | ATTCTGGTACTGCATCATCGACATTTTGCATTCCGACACCATTTATCATTTACTAACATT |
| CUST_46236_PI426222305 | JHI_St_60k_v1 | DMT400053238 | GATTTCGGAGGTGACATGAGAGAAAGTGAATTATATTGAGGGACGTAAGTTACTATTGAA |
| CUST_46240_PI426222305 | JHI_St_60k_v1 | DMT400053237 | CTAACATCTTGTTCATTTCTCTGTTTCTCAGGAGGTGACCTGAGAGAAAGTGAGTTATAT |
| CUST_46253_PI426222305 | JHI_St_60k_v1 | DMT400053236 | AGGGTTTAATTTCTAACTGCATTGTAACTGTGCAAGTGTTCTAATGAATGAAATCCCTTC |
| CUST_46246_PI426222305 | JHI_St_60k_v1 | DMT400053239 | CAAGCAGAACAAGCTGTTATCGATGCGCTTCCTTAAAAAGTGATTATTATGGTATTGGGG |
Microarray Signals from CUST_46246_PI426222305
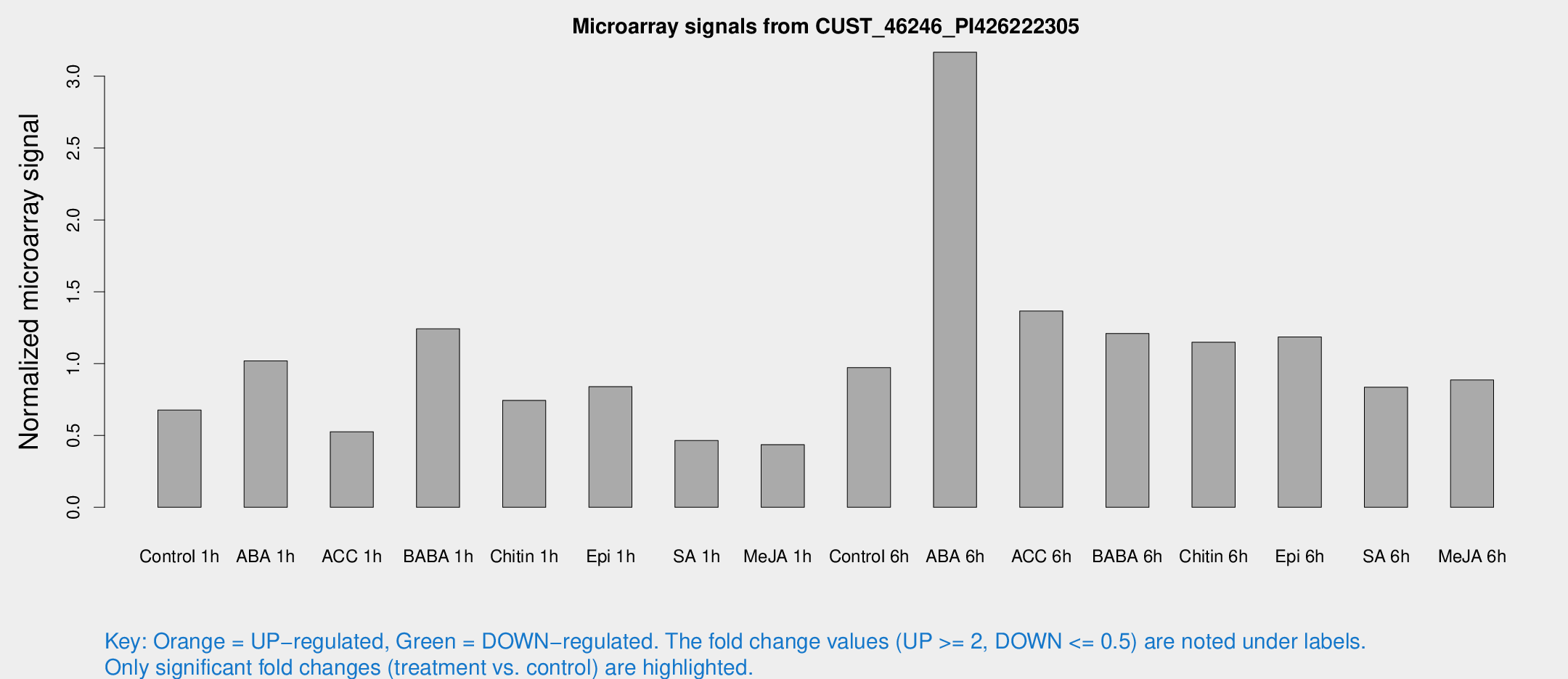
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.7348 | 17.016 | 0.676455 | 0.406487 |
| ABA 1h | 69.28 | 33.7968 | 1.0188 | 0.660442 |
| ACC 1h | 39.8921 | 15.2451 | 0.52586 | 0.32925 |
| BABA 1h | 73.346 | 20.2246 | 1.24224 | 0.251754 |
| Chitin 1h | 39.5897 | 8.66121 | 0.743945 | 0.102762 |
| Epi 1h | 42.992 | 8.23268 | 0.83937 | 0.213115 |
| SA 1h | 36.8742 | 14.7604 | 0.464504 | 0.364739 |
| Me-JA 1h | 21.7048 | 6.21304 | 0.435858 | 0.162371 |
| Control 6h | 55.9638 | 6.79651 | 0.972079 | 0.0885674 |
| ABA 6h | 192.813 | 19.398 | 3.16678 | 0.399725 |
| ACC 6h | 92.8792 | 19.9044 | 1.36542 | 0.174887 |
| BABA 6h | 85.3979 | 23.8598 | 1.20963 | 0.408245 |
| Chitin 6h | 70.026 | 7.17361 | 1.1489 | 0.145317 |
| Epi 6h | 78.6016 | 14.0164 | 1.18573 | 0.337713 |
| SA 6h | 56.9682 | 19.8939 | 0.835531 | 0.346873 |
| Me-JA 6h | 53.627 | 14.0161 | 0.885878 | 0.166248 |
Source Transcript PGSC0003DMT400053239 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26695.1 | +2 | 2e-62 | 194 | 84/134 (63%) | Ran BP2/NZF zinc finger-like superfamily protein | chr2:11365275-11365789 FORWARD LENGTH=138 |
