Probe CUST_46240_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46240_PI426222305 | JHI_St_60k_v1 | DMT400053237 | CTAACATCTTGTTCATTTCTCTGTTTCTCAGGAGGTGACCTGAGAGAAAGTGAGTTATAT |
All Microarray Probes Designed to Gene DMG401020664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46274_PI426222305 | JHI_St_60k_v1 | DMT400053235 | ATTCTGGTACTGCATCATCGACATTTTGCATTCCGACACCATTTATCATTTACTAACATT |
| CUST_46236_PI426222305 | JHI_St_60k_v1 | DMT400053238 | GATTTCGGAGGTGACATGAGAGAAAGTGAATTATATTGAGGGACGTAAGTTACTATTGAA |
| CUST_46240_PI426222305 | JHI_St_60k_v1 | DMT400053237 | CTAACATCTTGTTCATTTCTCTGTTTCTCAGGAGGTGACCTGAGAGAAAGTGAGTTATAT |
| CUST_46253_PI426222305 | JHI_St_60k_v1 | DMT400053236 | AGGGTTTAATTTCTAACTGCATTGTAACTGTGCAAGTGTTCTAATGAATGAAATCCCTTC |
| CUST_46246_PI426222305 | JHI_St_60k_v1 | DMT400053239 | CAAGCAGAACAAGCTGTTATCGATGCGCTTCCTTAAAAAGTGATTATTATGGTATTGGGG |
Microarray Signals from CUST_46240_PI426222305
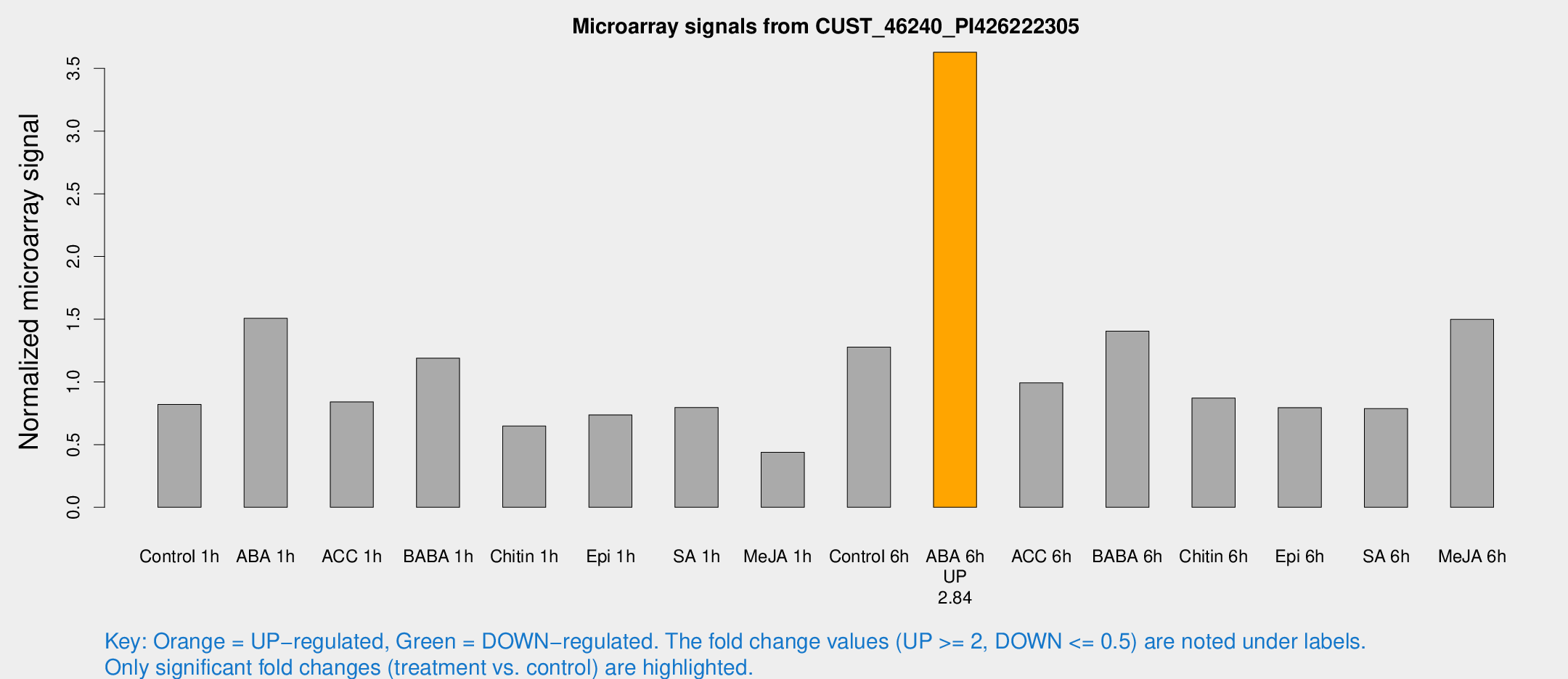
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.8323 | 3.16934 | 0.81997 | 0.27721 |
| ABA 1h | 30.9382 | 13.5396 | 1.50676 | 1.44212 |
| ACC 1h | 16.8591 | 7.13914 | 0.840204 | 0.383043 |
| BABA 1h | 20.3244 | 5.85529 | 1.18959 | 0.316327 |
| Chitin 1h | 9.94829 | 3.02195 | 0.647837 | 0.229764 |
| Epi 1h | 11.0415 | 3.02526 | 0.736797 | 0.261889 |
| SA 1h | 15.8443 | 6.00705 | 0.795677 | 0.412641 |
| Me-JA 1h | 5.65044 | 3.03095 | 0.439136 | 0.233663 |
| Control 6h | 20.816 | 3.39551 | 1.277 | 0.214079 |
| ABA 6h | 61.7368 | 5.98464 | 3.62886 | 0.337727 |
| ACC 6h | 18.5767 | 3.88263 | 0.992443 | 0.204651 |
| BABA 6h | 26.4395 | 6.11801 | 1.40467 | 0.320607 |
| Chitin 6h | 15.1662 | 3.49125 | 0.871682 | 0.219257 |
| Epi 6h | 15.9823 | 4.8818 | 0.795121 | 0.377062 |
| SA 6h | 12.6771 | 3.31873 | 0.786528 | 0.219679 |
| Me-JA 6h | 29.4969 | 13.8629 | 1.49813 | 0.648446 |
Source Transcript PGSC0003DMT400053237 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26695.1 | +2 | 3e-62 | 194 | 86/135 (64%) | Ran BP2/NZF zinc finger-like superfamily protein | chr2:11365275-11365789 FORWARD LENGTH=138 |
