Probe CUST_45942_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
All Microarray Probes Designed to Gene DMG400018659
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
Microarray Signals from CUST_45942_PI426222305
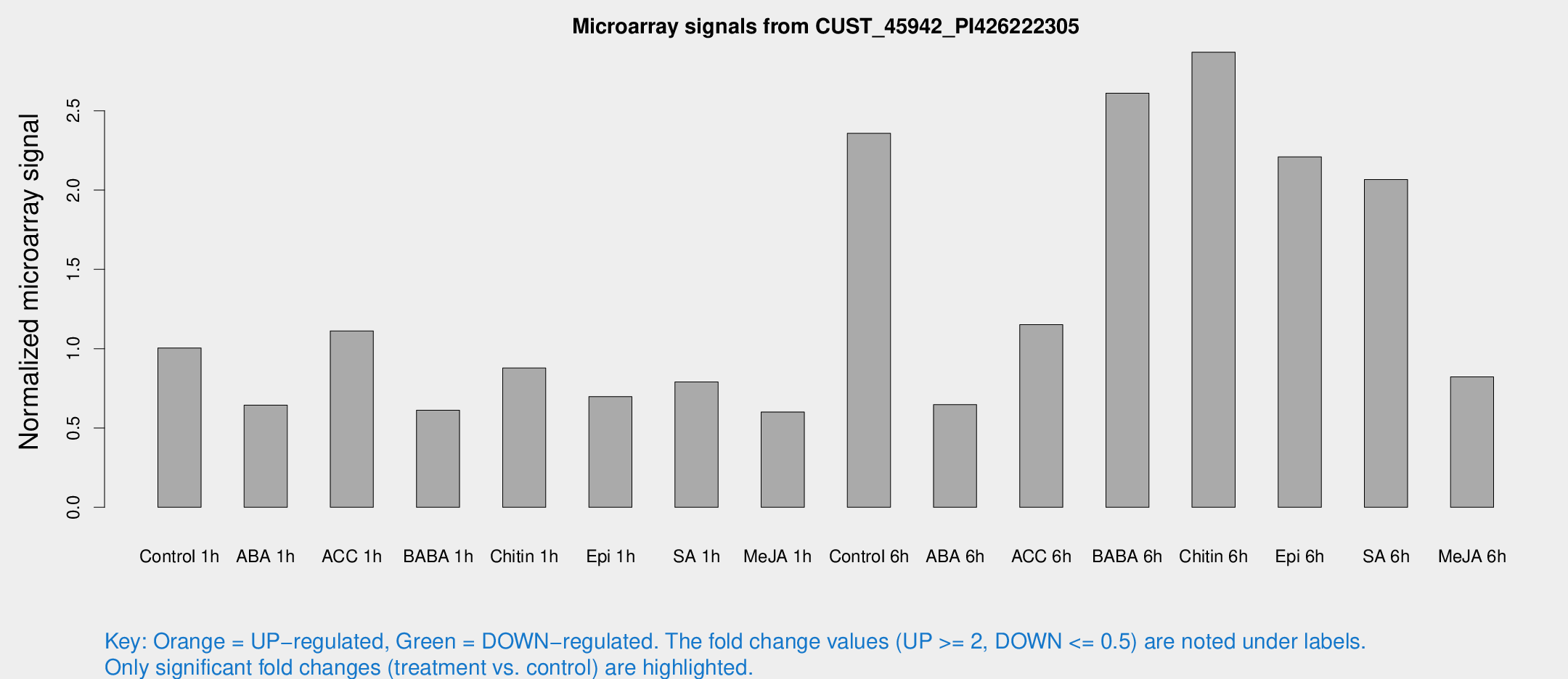
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 79.1859 | 16.1371 | 1.00478 | 0.163915 |
| ABA 1h | 43.8162 | 5.52415 | 0.644038 | 0.115322 |
| ACC 1h | 90.7182 | 18.6226 | 1.11222 | 0.165425 |
| BABA 1h | 46.0864 | 7.78111 | 0.611516 | 0.0634602 |
| Chitin 1h | 64.1926 | 17.2142 | 0.877927 | 0.289636 |
| Epi 1h | 46.2909 | 4.97625 | 0.697896 | 0.0664658 |
| SA 1h | 62.8911 | 9.62006 | 0.790316 | 0.159743 |
| Me-JA 1h | 37.573 | 4.10929 | 0.601092 | 0.0664277 |
| Control 6h | 195.147 | 49.7789 | 2.35792 | 0.533567 |
| ABA 6h | 53.7646 | 9.27795 | 0.647321 | 0.137693 |
| ACC 6h | 106.403 | 26.0839 | 1.15131 | 0.153115 |
| BABA 6h | 230.278 | 47.5265 | 2.61085 | 0.489245 |
| Chitin 6h | 233.912 | 23.8328 | 2.86904 | 0.228787 |
| Epi 6h | 189.338 | 11.7016 | 2.20952 | 0.296701 |
| SA 6h | 156.406 | 16.1219 | 2.06669 | 0.192679 |
| Me-JA 6h | 75.5171 | 31.1297 | 0.822111 | 0.344937 |
Source Transcript PGSC0003DMT400048021 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19250.1 | +1 | 9e-93 | 173 | 88/199 (44%) | flavin-dependent monooxygenase 1 | chr1:6650656-6653053 REVERSE LENGTH=530 |
