Probe CUST_45900_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
All Microarray Probes Designed to Gene DMG400018659
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
Microarray Signals from CUST_45900_PI426222305
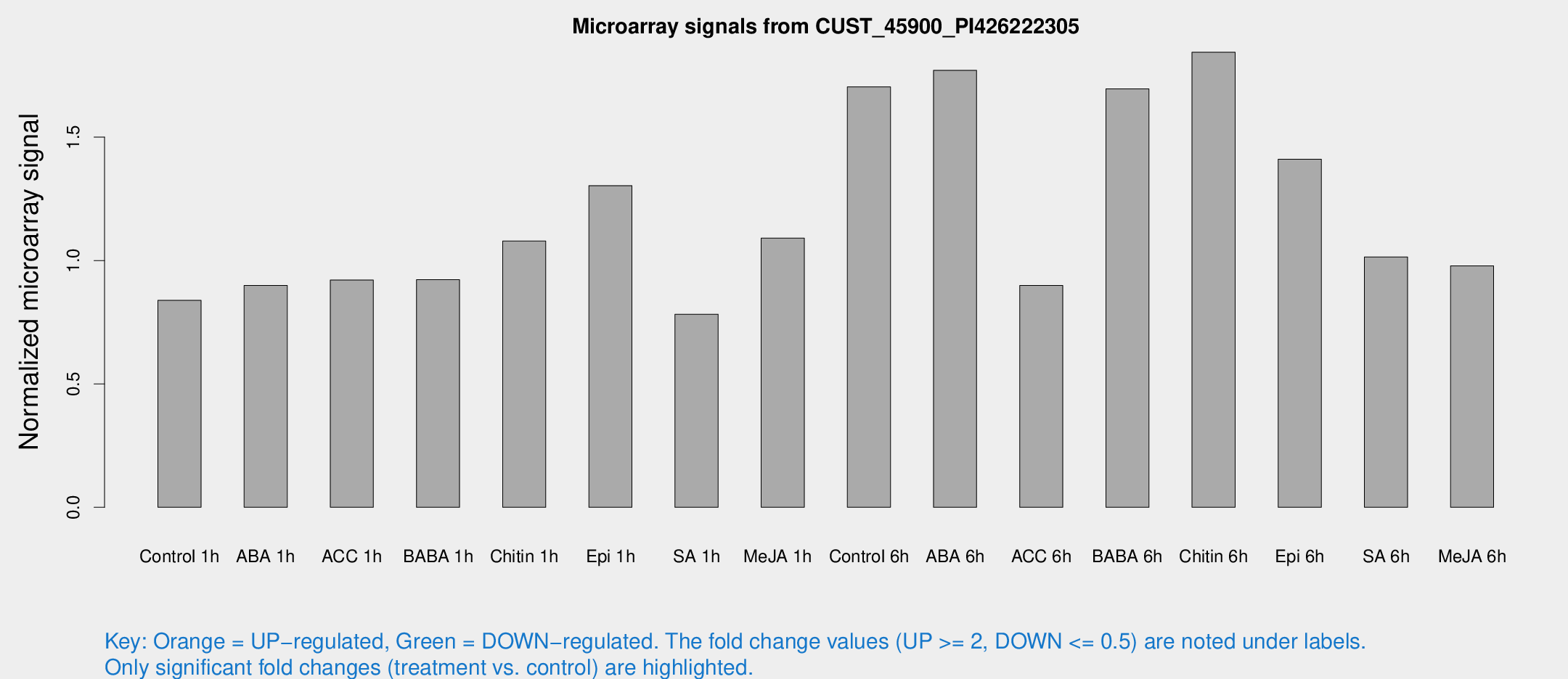
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.72839 | 3.32053 | 0.838565 | 0.485781 |
| ABA 1h | 5.43169 | 3.14615 | 0.899179 | 0.520707 |
| ACC 1h | 6.48054 | 3.50759 | 0.920732 | 0.502095 |
| BABA 1h | 6.07542 | 3.52223 | 0.92228 | 0.534155 |
| Chitin 1h | 7.03078 | 3.34312 | 1.07898 | 0.550916 |
| Epi 1h | 8.65529 | 3.29299 | 1.30342 | 0.616495 |
| SA 1h | 5.4965 | 3.18837 | 0.782523 | 0.453234 |
| Me-JA 1h | 6.10777 | 3.37246 | 1.09106 | 0.601338 |
| Control 6h | 12.8276 | 3.81026 | 1.70384 | 0.633028 |
| ABA 6h | 14.5933 | 5.01394 | 1.77084 | 0.794338 |
| ACC 6h | 7.17609 | 4.25356 | 0.898712 | 0.521969 |
| BABA 6h | 14.5729 | 4.89862 | 1.69566 | 0.618201 |
| Chitin 6h | 14.7436 | 4.30359 | 1.84407 | 0.647724 |
| Epi 6h | 13.1513 | 5.89607 | 1.41074 | 0.651473 |
| SA 6h | 6.87104 | 3.85174 | 1.01452 | 0.570507 |
| Me-JA 6h | 6.6842 | 3.61589 | 0.978677 | 0.532116 |
Source Transcript PGSC0003DMT400048022 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19250.1 | +3 | 1e-87 | 178 | 114/319 (36%) | flavin-dependent monooxygenase 1 | chr1:6650656-6653053 REVERSE LENGTH=530 |
