Probe CUST_45912_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
All Microarray Probes Designed to Gene DMG400018659
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
Microarray Signals from CUST_45912_PI426222305
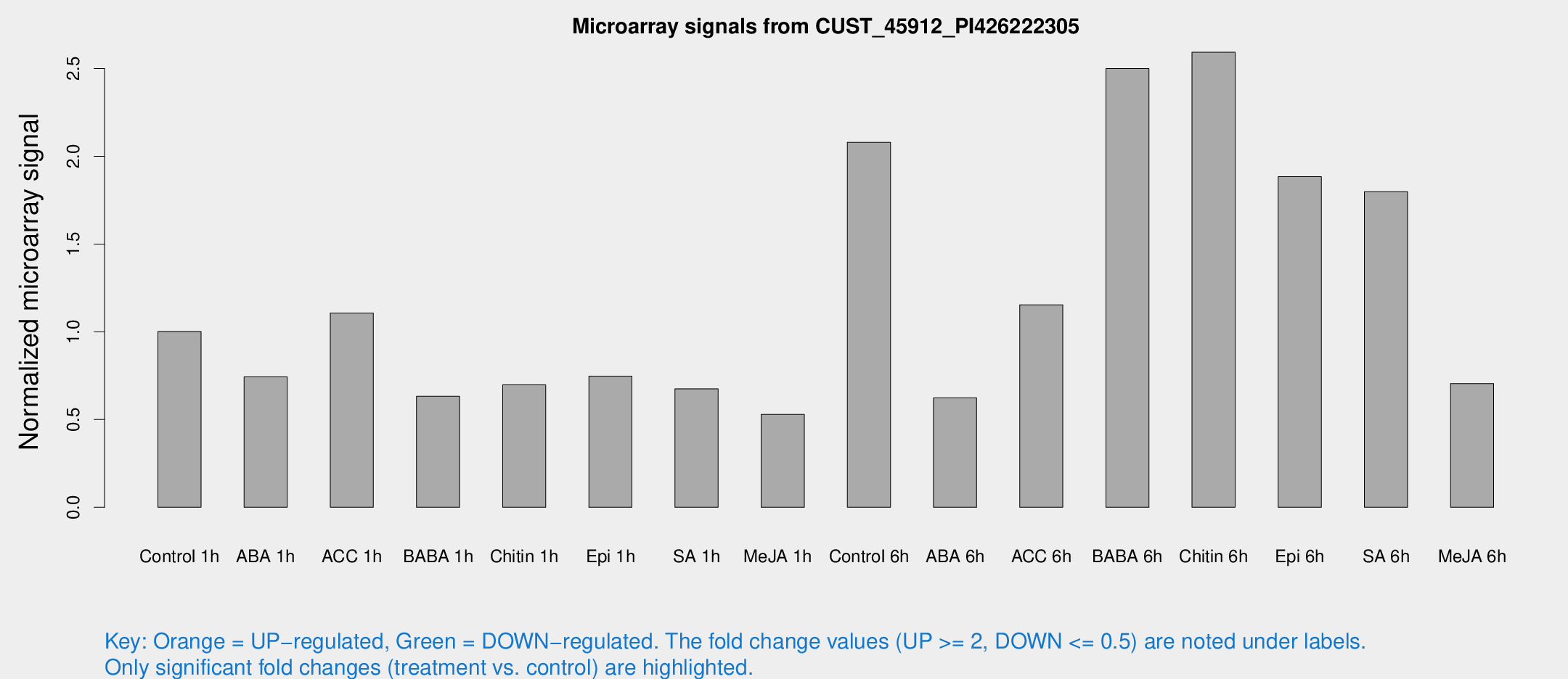
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 86.0667 | 13.6676 | 1.00142 | 0.0998176 |
| ABA 1h | 56.2736 | 8.25756 | 0.743379 | 0.144438 |
| ACC 1h | 98.4242 | 18.7406 | 1.10679 | 0.145726 |
| BABA 1h | 51.4015 | 4.93771 | 0.632544 | 0.0607352 |
| Chitin 1h | 55.9074 | 13.114 | 0.697646 | 0.239071 |
| Epi 1h | 55.4456 | 8.47554 | 0.747163 | 0.115982 |
| SA 1h | 59.0949 | 7.62943 | 0.67548 | 0.116824 |
| Me-JA 1h | 36.3861 | 4.37197 | 0.529182 | 0.0787452 |
| Control 6h | 195.944 | 56.4532 | 2.07929 | 0.604258 |
| ABA 6h | 56.7999 | 8.9886 | 0.623486 | 0.13076 |
| ACC 6h | 120.944 | 36.1833 | 1.1537 | 0.193754 |
| BABA 6h | 240.64 | 41.2633 | 2.49965 | 0.402681 |
| Chitin 6h | 233.871 | 26.646 | 2.59292 | 0.209479 |
| Epi 6h | 177.814 | 11.2036 | 1.88381 | 0.133683 |
| SA 6h | 149.921 | 12.341 | 1.79849 | 0.115498 |
| Me-JA 6h | 75.2961 | 30.7154 | 0.705495 | 0.387542 |
Source Transcript PGSC0003DMT400048020 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19250.1 | +1 | 4e-116 | 360 | 196/497 (39%) | flavin-dependent monooxygenase 1 | chr1:6650656-6653053 REVERSE LENGTH=530 |
