Probe CUST_45941_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
All Microarray Probes Designed to Gene DMG400018659
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
Microarray Signals from CUST_45941_PI426222305
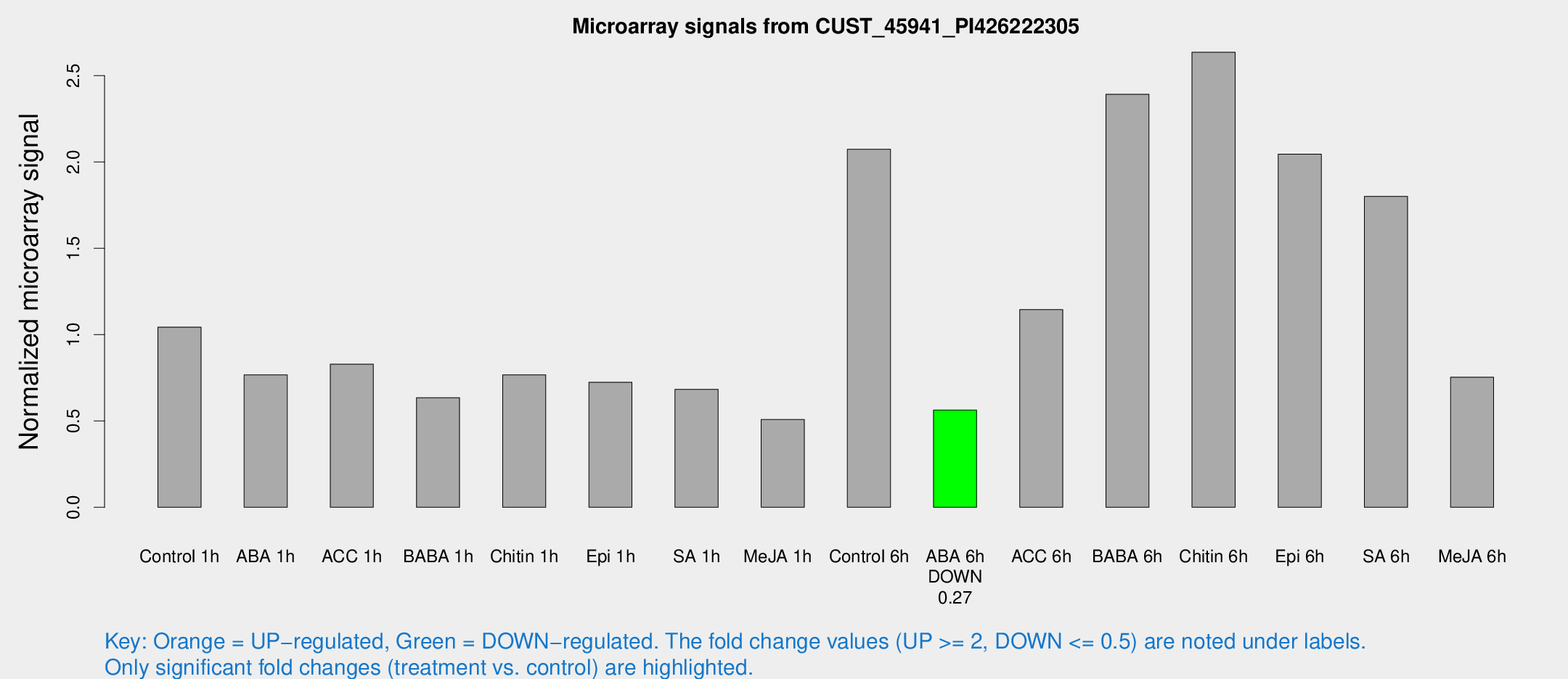
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 90.0415 | 16.3722 | 1.04342 | 0.12275 |
| ABA 1h | 58.4002 | 10.1509 | 0.766979 | 0.178246 |
| ACC 1h | 75.2068 | 18.3715 | 0.828698 | 0.158519 |
| BABA 1h | 51.5108 | 4.59824 | 0.634828 | 0.0581364 |
| Chitin 1h | 60.9769 | 14.7171 | 0.767542 | 0.206441 |
| Epi 1h | 52.5154 | 4.47653 | 0.724555 | 0.0618266 |
| SA 1h | 59.2358 | 5.38754 | 0.683435 | 0.101775 |
| Me-JA 1h | 35.7207 | 5.77808 | 0.508587 | 0.0709433 |
| Control 6h | 197.604 | 62.3974 | 2.07355 | 0.649227 |
| ABA 6h | 50.3802 | 4.58612 | 0.562819 | 0.0661603 |
| ACC 6h | 114.168 | 22.5592 | 1.14509 | 0.0782498 |
| BABA 6h | 229.968 | 37.9988 | 2.39254 | 0.387247 |
| Chitin 6h | 236.21 | 18.0151 | 2.63573 | 0.15803 |
| Epi 6h | 193.188 | 11.826 | 2.04582 | 0.164915 |
| SA 6h | 149.602 | 9.36193 | 1.80093 | 0.15229 |
| Me-JA 6h | 79.9147 | 31.8666 | 0.753942 | 0.406906 |
Source Transcript PGSC0003DMT400048019 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19250.1 | +2 | 1e-71 | 166 | 92/241 (38%) | flavin-dependent monooxygenase 1 | chr1:6650656-6653053 REVERSE LENGTH=530 |
