Probe CUST_42748_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42748_PI426222305 | JHI_St_60k_v1 | DMT400052383 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
All Microarray Probes Designed to Gene DMG400020335
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42660_PI426222305 | JHI_St_60k_v1 | DMT400052388 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42708_PI426222305 | JHI_St_60k_v1 | DMT400052384 | CAGGTATGCTTATCATCCTCAATGCTTTACATCTTCCCATTTCATCACTTAAATCAAACT |
| CUST_42704_PI426222305 | JHI_St_60k_v1 | DMT400052385 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42718_PI426222305 | JHI_St_60k_v1 | DMT400052389 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
| CUST_42748_PI426222305 | JHI_St_60k_v1 | DMT400052383 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
Microarray Signals from CUST_42748_PI426222305
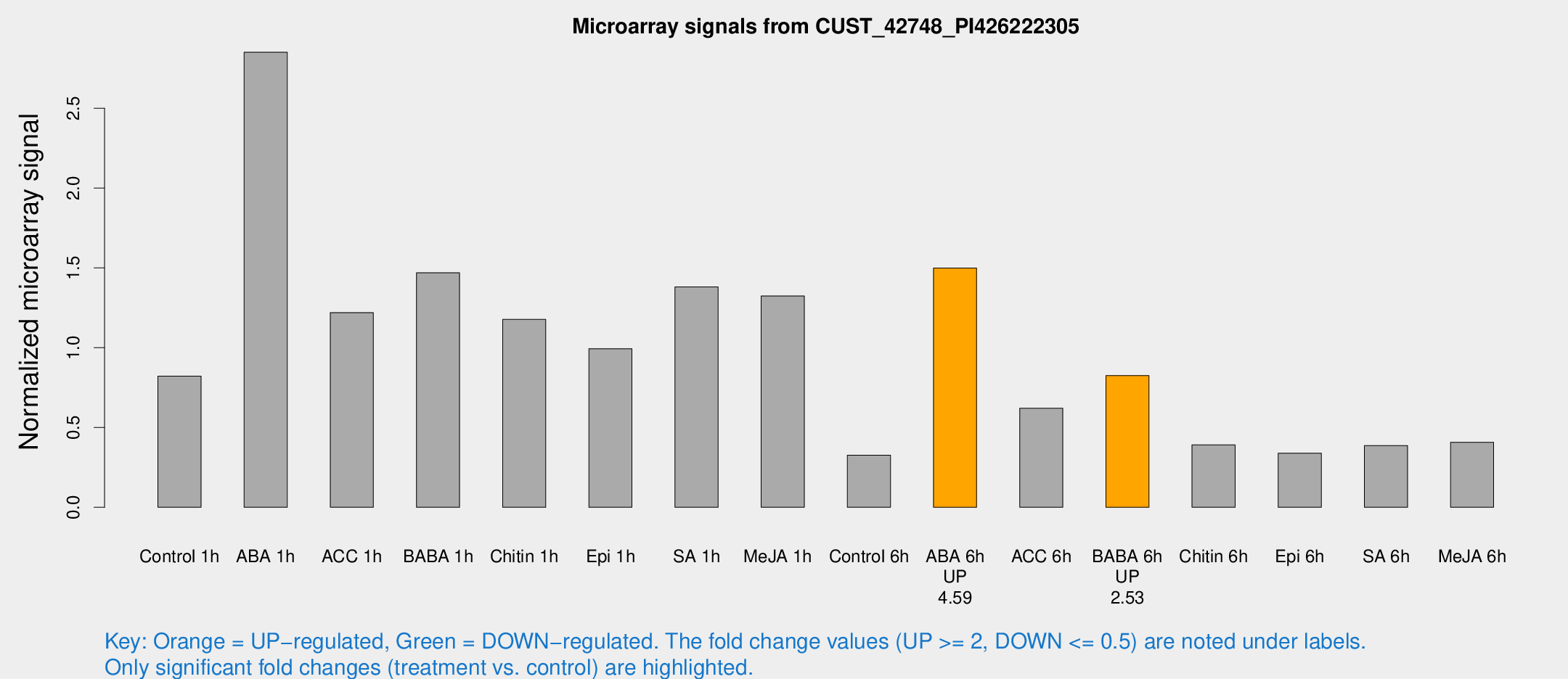
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 125.688 | 26.4851 | 0.821699 | 0.134133 |
| ABA 1h | 380.667 | 66.7188 | 2.85137 | 0.51711 |
| ACC 1h | 189.373 | 33.6598 | 1.22007 | 0.168401 |
| BABA 1h | 208.292 | 12.5696 | 1.46944 | 0.097865 |
| Chitin 1h | 158.154 | 20.2298 | 1.1776 | 0.109459 |
| Epi 1h | 126.381 | 8.01718 | 0.993863 | 0.0629937 |
| SA 1h | 215.288 | 38.2119 | 1.38083 | 0.176317 |
| Me-JA 1h | 158.904 | 9.81705 | 1.32404 | 0.0816024 |
| Control 6h | 53.2057 | 15.1341 | 0.32649 | 0.0830796 |
| ABA 6h | 241.526 | 45.1073 | 1.49875 | 0.18822 |
| ACC 6h | 105.309 | 9.72538 | 0.620632 | 0.0434401 |
| BABA 6h | 140.138 | 25.409 | 0.825691 | 0.119599 |
| Chitin 6h | 61.1993 | 5.39991 | 0.39148 | 0.0346604 |
| Epi 6h | 56.8779 | 6.14608 | 0.3395 | 0.0316656 |
| SA 6h | 57.0516 | 7.36496 | 0.38691 | 0.0597312 |
| Me-JA 6h | 63.2173 | 15.901 | 0.407623 | 0.0849333 |
Source Transcript PGSC0003DMT400052383 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37980.1 | +3 | 0.0 | 731 | 392/648 (60%) | O-fucosyltransferase family protein | chr2:15894162-15897452 REVERSE LENGTH=638 |
