Probe CUST_42718_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42718_PI426222305 | JHI_St_60k_v1 | DMT400052389 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
All Microarray Probes Designed to Gene DMG400020335
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42660_PI426222305 | JHI_St_60k_v1 | DMT400052388 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42708_PI426222305 | JHI_St_60k_v1 | DMT400052384 | CAGGTATGCTTATCATCCTCAATGCTTTACATCTTCCCATTTCATCACTTAAATCAAACT |
| CUST_42704_PI426222305 | JHI_St_60k_v1 | DMT400052385 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42718_PI426222305 | JHI_St_60k_v1 | DMT400052389 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
| CUST_42748_PI426222305 | JHI_St_60k_v1 | DMT400052383 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
Microarray Signals from CUST_42718_PI426222305
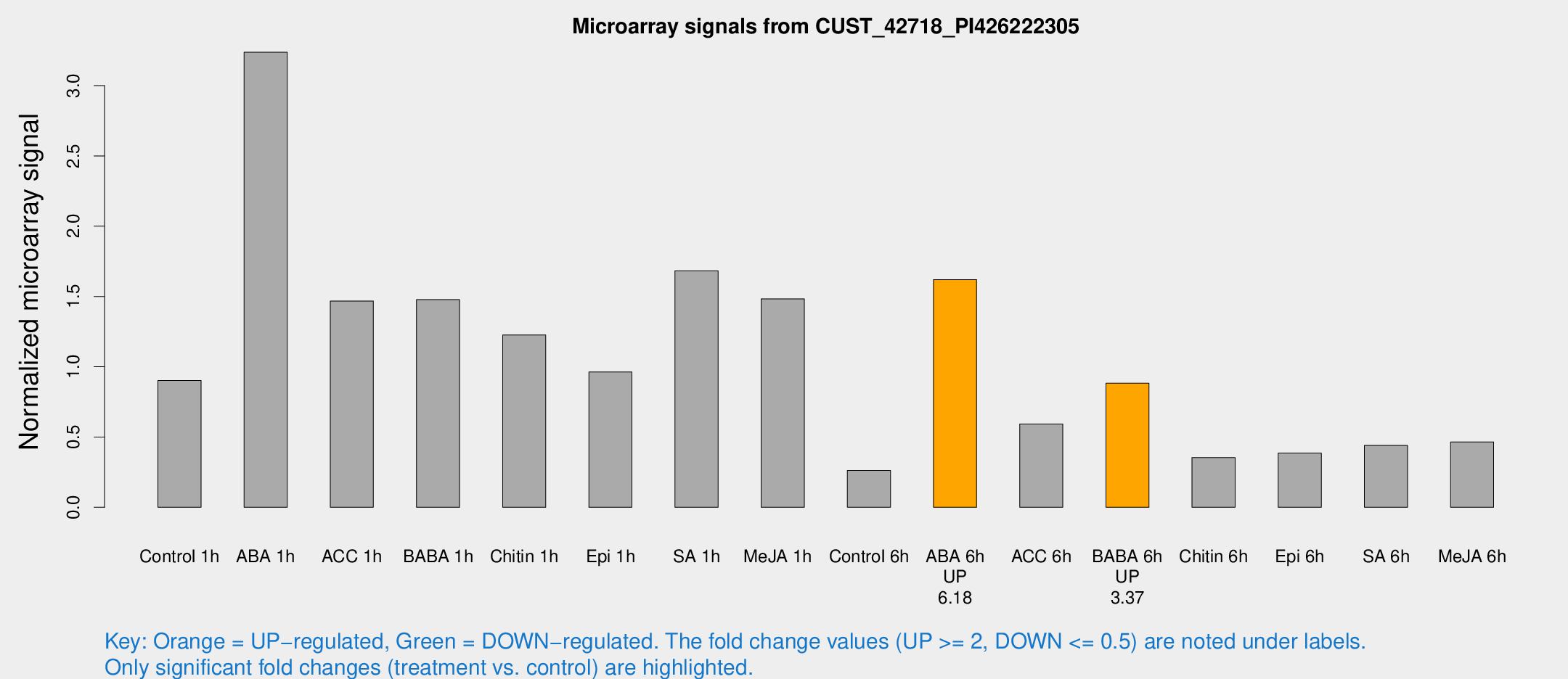
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 124.109 | 28.8607 | 0.902067 | 0.152757 |
| ABA 1h | 386.155 | 67.9724 | 3.23734 | 0.557487 |
| ACC 1h | 199.707 | 22.5004 | 1.46702 | 0.128834 |
| BABA 1h | 186.952 | 11.3181 | 1.47777 | 0.126 |
| Chitin 1h | 147.625 | 21.8352 | 1.22545 | 0.154964 |
| Epi 1h | 109.538 | 7.14265 | 0.962295 | 0.0637651 |
| SA 1h | 231.03 | 31.7524 | 1.68234 | 0.14764 |
| Me-JA 1h | 158.971 | 9.74668 | 1.48278 | 0.141269 |
| Control 6h | 38.9368 | 13.53 | 0.261975 | 0.0747068 |
| ABA 6h | 227.433 | 21.1216 | 1.62006 | 0.0966318 |
| ACC 6h | 89.6896 | 7.57483 | 0.592801 | 0.0496266 |
| BABA 6h | 139.164 | 37.3022 | 0.882384 | 0.24176 |
| Chitin 6h | 49.9101 | 5.45593 | 0.353778 | 0.0460147 |
| Epi 6h | 57.5391 | 5.80332 | 0.385535 | 0.0333637 |
| SA 6h | 58.3767 | 8.87528 | 0.440689 | 0.0404373 |
| Me-JA 6h | 63.2738 | 13.6717 | 0.464913 | 0.0702602 |
Source Transcript PGSC0003DMT400052389 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37980.1 | +3 | 2e-125 | 379 | 179/232 (77%) | O-fucosyltransferase family protein | chr2:15894162-15897452 REVERSE LENGTH=638 |
