Probe CUST_42660_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42660_PI426222305 | JHI_St_60k_v1 | DMT400052388 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
All Microarray Probes Designed to Gene DMG400020335
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42660_PI426222305 | JHI_St_60k_v1 | DMT400052388 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42708_PI426222305 | JHI_St_60k_v1 | DMT400052384 | CAGGTATGCTTATCATCCTCAATGCTTTACATCTTCCCATTTCATCACTTAAATCAAACT |
| CUST_42704_PI426222305 | JHI_St_60k_v1 | DMT400052385 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42718_PI426222305 | JHI_St_60k_v1 | DMT400052389 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
| CUST_42748_PI426222305 | JHI_St_60k_v1 | DMT400052383 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
Microarray Signals from CUST_42660_PI426222305
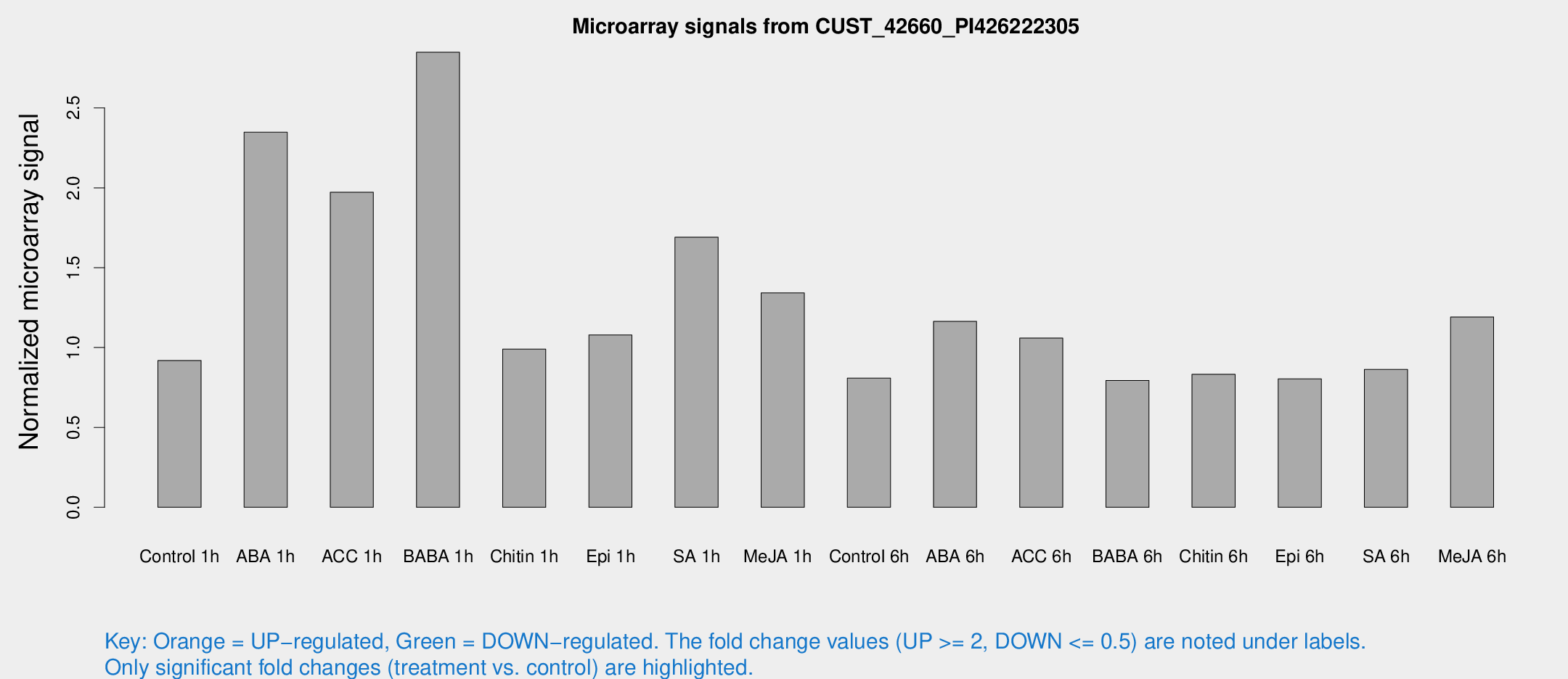
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.3414 | 3.44268 | 0.919276 | 0.4645 |
| ABA 1h | 19.8625 | 8.8953 | 2.34755 | 1.30593 |
| ACC 1h | 15.739 | 4.39703 | 1.97178 | 0.580932 |
| BABA 1h | 22.7661 | 6.76865 | 2.84862 | 1.26731 |
| Chitin 1h | 6.90654 | 3.43344 | 0.989752 | 0.498535 |
| Epi 1h | 7.38831 | 3.3312 | 1.07907 | 0.516896 |
| SA 1h | 13.9569 | 3.51243 | 1.69115 | 0.483479 |
| Me-JA 1h | 9.03837 | 3.54417 | 1.34218 | 0.6251 |
| Control 6h | 6.16472 | 3.57743 | 0.808078 | 0.468044 |
| ABA 6h | 10.3877 | 3.72196 | 1.16391 | 0.501844 |
| ACC 6h | 9.74667 | 4.20539 | 1.05951 | 0.50475 |
| BABA 6h | 6.76997 | 3.9317 | 0.794244 | 0.46011 |
| Chitin 6h | 6.74451 | 3.91273 | 0.832868 | 0.482498 |
| Epi 6h | 6.99523 | 4.11496 | 0.804416 | 0.465811 |
| SA 6h | 6.48376 | 3.75537 | 0.862703 | 0.499574 |
| Me-JA 6h | 9.51565 | 3.55882 | 1.19107 | 0.49798 |
Source Transcript PGSC0003DMT400052388 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37980.1 | +2 | 0.0 | 627 | 300/377 (80%) | O-fucosyltransferase family protein | chr2:15894162-15897452 REVERSE LENGTH=638 |
