Probe CUST_42704_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42704_PI426222305 | JHI_St_60k_v1 | DMT400052385 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
All Microarray Probes Designed to Gene DMG400020335
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42660_PI426222305 | JHI_St_60k_v1 | DMT400052388 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42708_PI426222305 | JHI_St_60k_v1 | DMT400052384 | CAGGTATGCTTATCATCCTCAATGCTTTACATCTTCCCATTTCATCACTTAAATCAAACT |
| CUST_42704_PI426222305 | JHI_St_60k_v1 | DMT400052385 | CAGATGCAGATTTGCTCTTCCTAGACAAATGTTTGTGTTAGCCTTTCATTTTTAGCTTAA |
| CUST_42718_PI426222305 | JHI_St_60k_v1 | DMT400052389 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
| CUST_42748_PI426222305 | JHI_St_60k_v1 | DMT400052383 | GTTGTAACAAGCAAAGCAGTACTTGGAAGCAGAGATGATGTTTTATAAGATGGACAAAAG |
Microarray Signals from CUST_42704_PI426222305
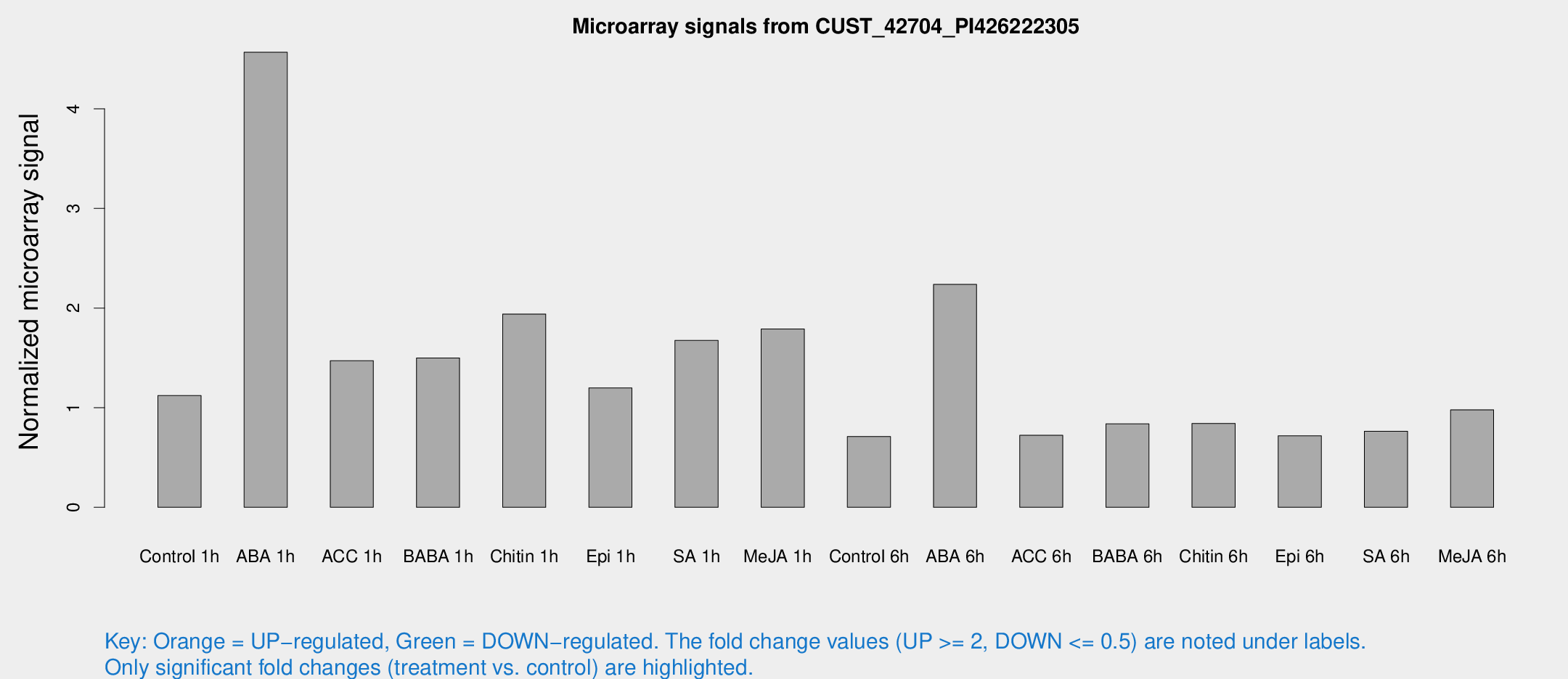
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.03885 | 3.4173 | 1.12098 | 0.453541 |
| ABA 1h | 29.1259 | 3.2251 | 4.5677 | 0.50671 |
| ACC 1h | 11.9484 | 3.22114 | 1.47094 | 0.516346 |
| BABA 1h | 11.6224 | 3.29543 | 1.49911 | 0.529806 |
| Chitin 1h | 14.5421 | 5.82655 | 1.94003 | 0.795463 |
| Epi 1h | 8.6806 | 3.56546 | 1.19817 | 0.551204 |
| SA 1h | 13.8986 | 4.54957 | 1.67484 | 0.570672 |
| Me-JA 1h | 12.4817 | 5.16564 | 1.79047 | 0.800798 |
| Control 6h | 5.12874 | 2.97673 | 0.710557 | 0.412362 |
| ABA 6h | 18.5021 | 4.83622 | 2.23688 | 0.608445 |
| ACC 6h | 6.0953 | 3.60948 | 0.722516 | 0.418488 |
| BABA 6h | 7.05404 | 3.37195 | 0.837747 | 0.427121 |
| Chitin 6h | 6.54868 | 3.30172 | 0.842401 | 0.438014 |
| Epi 6h | 5.86729 | 3.42687 | 0.717192 | 0.415737 |
| SA 6h | 5.43713 | 3.15072 | 0.762879 | 0.441777 |
| Me-JA 6h | 7.70647 | 3.0151 | 0.978539 | 0.460265 |
Source Transcript PGSC0003DMT400052385 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37980.1 | +3 | 0.0 | 731 | 392/648 (60%) | O-fucosyltransferase family protein | chr2:15894162-15897452 REVERSE LENGTH=638 |
