Probe CUST_37071_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37071_PI426222305 | JHI_St_60k_v1 | DMT400061953 | GCGTTTTGTTCTATTATTAGAACGTGACACTCGCAGTAATGTTATTATGCAGGCACTTGA |
All Microarray Probes Designed to Gene DMG400024109
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37050_PI426222305 | JHI_St_60k_v1 | DMT400061949 | ATTCTGTCTGCACTTGACTTAGTTCCAGAGAAGATACACGAACGCTCTCCTATATTTCTT |
| CUST_37042_PI426222305 | JHI_St_60k_v1 | DMT400061950 | CAAGTACGCTTATTGGAAAGCAATAGCTAATAAGTTTGTTTCCCATGTTCATGATATCCT |
| CUST_37033_PI426222305 | JHI_St_60k_v1 | DMT400061951 | GCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTTTTT |
| CUST_37071_PI426222305 | JHI_St_60k_v1 | DMT400061953 | GCGTTTTGTTCTATTATTAGAACGTGACACTCGCAGTAATGTTATTATGCAGGCACTTGA |
| CUST_37028_PI426222305 | JHI_St_60k_v1 | DMT400061952 | GTTGCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTT |
Microarray Signals from CUST_37071_PI426222305
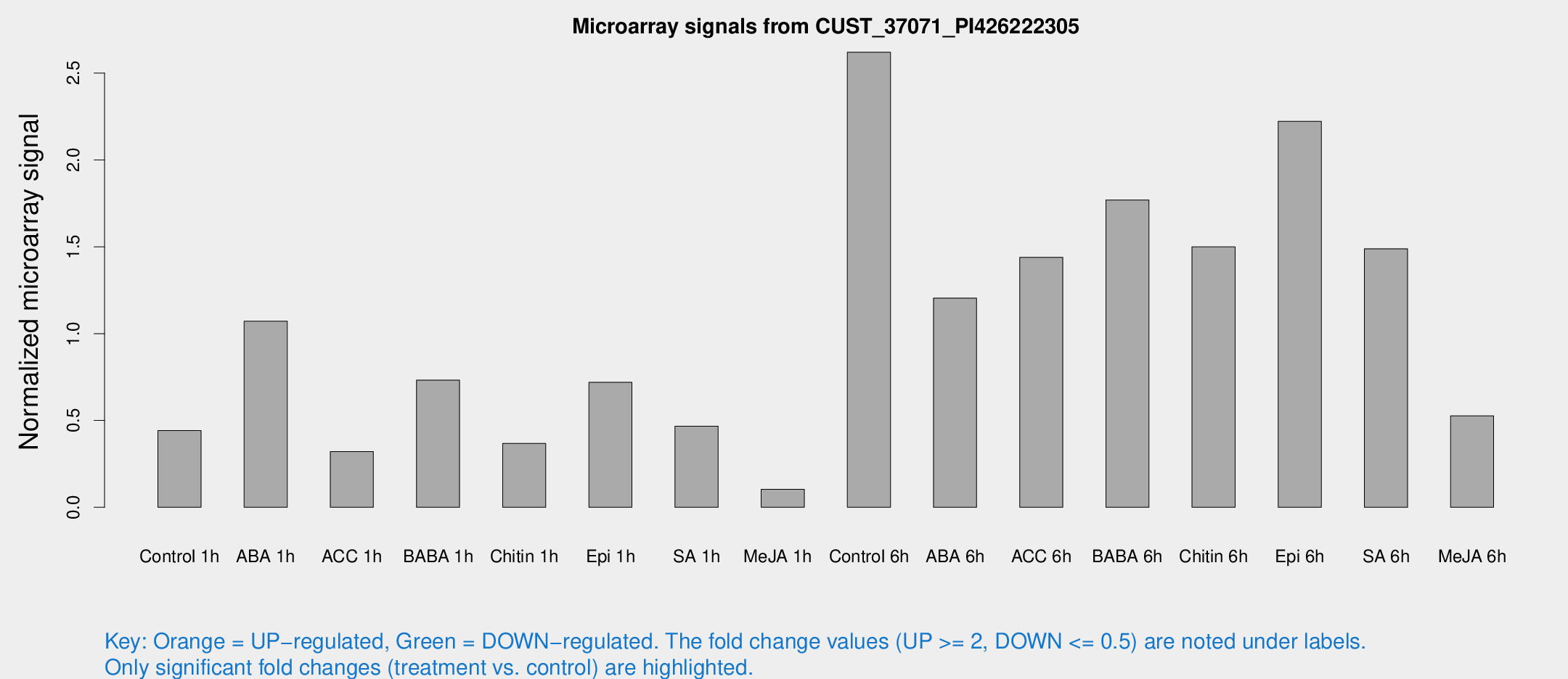
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.9428 | 12.6836 | 0.442196 | 0.107968 |
| ABA 1h | 94.8363 | 18.9313 | 1.07143 | 0.146603 |
| ACC 1h | 37.5742 | 12.8962 | 0.320589 | 0.130269 |
| BABA 1h | 75.8806 | 21.4243 | 0.732164 | 0.189955 |
| Chitin 1h | 33.7657 | 8.37243 | 0.36792 | 0.0669577 |
| Epi 1h | 61.2194 | 8.65405 | 0.719368 | 0.080784 |
| SA 1h | 52.3076 | 17.9213 | 0.466597 | 0.167297 |
| Me-JA 1h | 9.53633 | 3.91559 | 0.103817 | 0.0457548 |
| Control 6h | 275.343 | 74.6347 | 2.6199 | 0.597916 |
| ABA 6h | 124.37 | 9.97487 | 1.20453 | 0.0765857 |
| ACC 6h | 170.548 | 40.6474 | 1.43927 | 0.275202 |
| BABA 6h | 199.307 | 42.1622 | 1.76931 | 0.338932 |
| Chitin 6h | 157.504 | 23.5922 | 1.49955 | 0.180691 |
| Epi 6h | 252.058 | 53.2231 | 2.22236 | 0.425762 |
| SA 6h | 158.308 | 54.8349 | 1.48829 | 0.413141 |
| Me-JA 6h | 76.6645 | 36.633 | 0.526759 | 0.505085 |
Source Transcript PGSC0003DMT400061953 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G43670.1 | +2 | 6e-39 | 111 | 53/55 (96%) | Inositol monophosphatase family protein | chr1:16468184-16470347 FORWARD LENGTH=341 |
