Probe CUST_37033_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37033_PI426222305 | JHI_St_60k_v1 | DMT400061951 | GCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTTTTT |
All Microarray Probes Designed to Gene DMG400024109
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37050_PI426222305 | JHI_St_60k_v1 | DMT400061949 | ATTCTGTCTGCACTTGACTTAGTTCCAGAGAAGATACACGAACGCTCTCCTATATTTCTT |
| CUST_37042_PI426222305 | JHI_St_60k_v1 | DMT400061950 | CAAGTACGCTTATTGGAAAGCAATAGCTAATAAGTTTGTTTCCCATGTTCATGATATCCT |
| CUST_37033_PI426222305 | JHI_St_60k_v1 | DMT400061951 | GCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTTTTT |
| CUST_37071_PI426222305 | JHI_St_60k_v1 | DMT400061953 | GCGTTTTGTTCTATTATTAGAACGTGACACTCGCAGTAATGTTATTATGCAGGCACTTGA |
| CUST_37028_PI426222305 | JHI_St_60k_v1 | DMT400061952 | GTTGCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTT |
Microarray Signals from CUST_37033_PI426222305
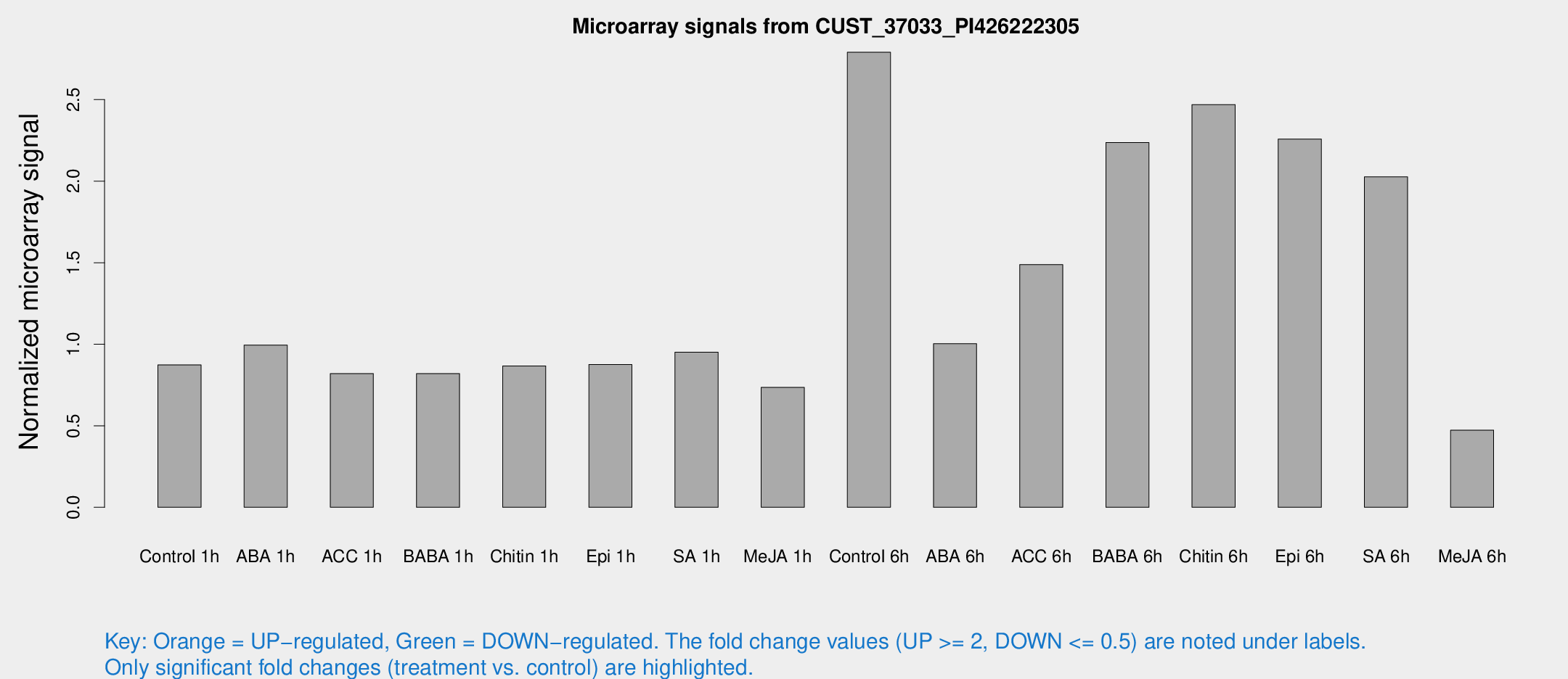
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 15289.4 | 1308.12 | 0.872856 | 0.0503948 |
| ABA 1h | 15587.2 | 2139.73 | 0.99513 | 0.0648152 |
| ACC 1h | 14986.5 | 2189.02 | 0.819567 | 0.0644894 |
| BABA 1h | 14986.6 | 3828.11 | 0.820006 | 0.175403 |
| Chitin 1h | 13635.3 | 1052.43 | 0.867073 | 0.0849345 |
| Epi 1h | 13205.2 | 764.103 | 0.875524 | 0.0505489 |
| SA 1h | 17429.3 | 2875.72 | 0.951501 | 0.100689 |
| Me-JA 1h | 10663.9 | 1601.39 | 0.735367 | 0.0478528 |
| Control 6h | 50833.4 | 10089.2 | 2.79073 | 0.384554 |
| ABA 6h | 18554.8 | 1072.01 | 1.00301 | 0.0750438 |
| ACC 6h | 31268.5 | 6876.01 | 1.48851 | 0.188047 |
| BABA 6h | 45029.3 | 8535.73 | 2.23691 | 0.425329 |
| Chitin 6h | 46848 | 7611.87 | 2.46981 | 0.31874 |
| Epi 6h | 44852.8 | 5260.31 | 2.25833 | 0.285418 |
| SA 6h | 35310.7 | 3910.45 | 2.02643 | 0.116996 |
| Me-JA 6h | 10632.2 | 5037.52 | 0.472821 | 0.246935 |
Source Transcript PGSC0003DMT400061951 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G43670.1 | +2 | 0.0 | 581 | 294/339 (87%) | Inositol monophosphatase family protein | chr1:16468184-16470347 FORWARD LENGTH=341 |
