Probe CUST_37042_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37042_PI426222305 | JHI_St_60k_v1 | DMT400061950 | CAAGTACGCTTATTGGAAAGCAATAGCTAATAAGTTTGTTTCCCATGTTCATGATATCCT |
All Microarray Probes Designed to Gene DMG400024109
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37050_PI426222305 | JHI_St_60k_v1 | DMT400061949 | ATTCTGTCTGCACTTGACTTAGTTCCAGAGAAGATACACGAACGCTCTCCTATATTTCTT |
| CUST_37042_PI426222305 | JHI_St_60k_v1 | DMT400061950 | CAAGTACGCTTATTGGAAAGCAATAGCTAATAAGTTTGTTTCCCATGTTCATGATATCCT |
| CUST_37033_PI426222305 | JHI_St_60k_v1 | DMT400061951 | GCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTTTTT |
| CUST_37071_PI426222305 | JHI_St_60k_v1 | DMT400061953 | GCGTTTTGTTCTATTATTAGAACGTGACACTCGCAGTAATGTTATTATGCAGGCACTTGA |
| CUST_37028_PI426222305 | JHI_St_60k_v1 | DMT400061952 | GTTGCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTT |
Microarray Signals from CUST_37042_PI426222305
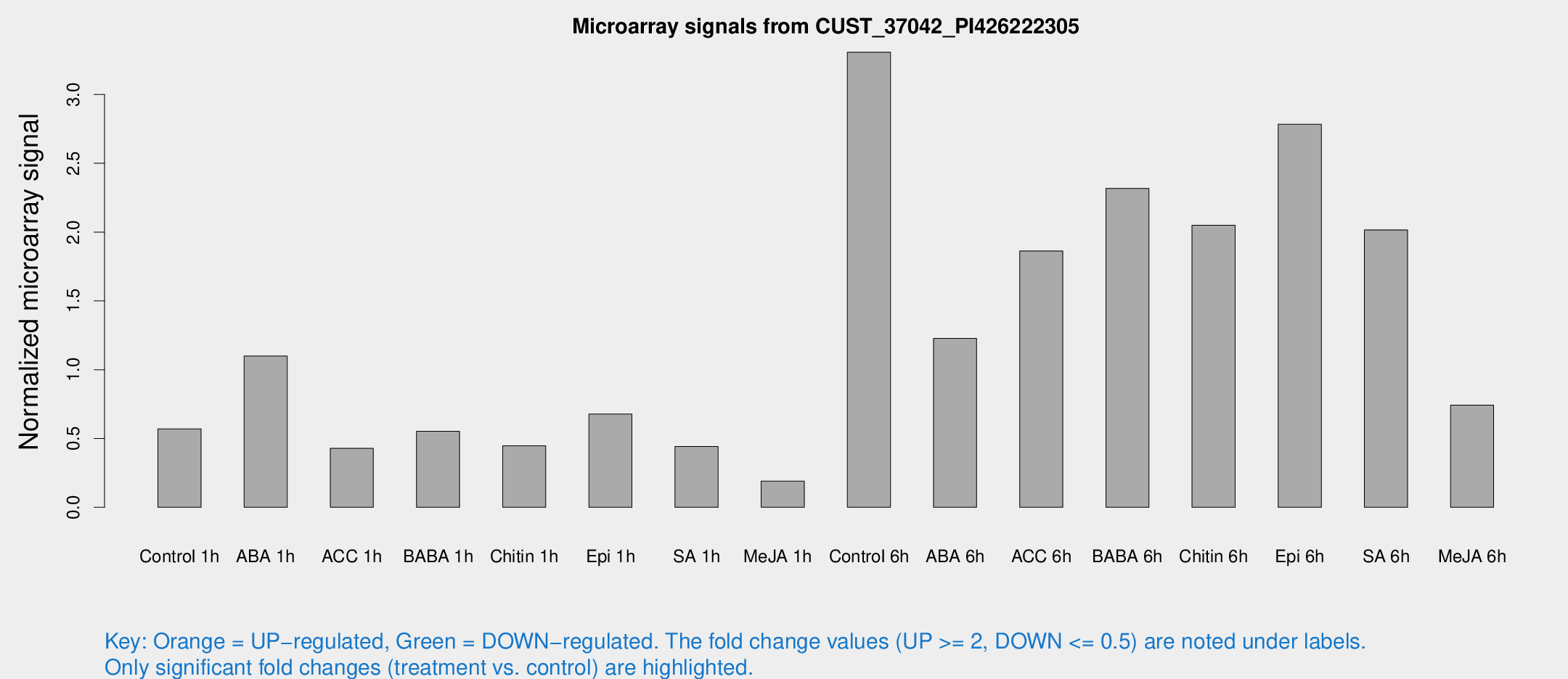
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 121.823 | 25.8626 | 0.570413 | 0.0863205 |
| ABA 1h | 207.233 | 40.3158 | 1.09911 | 0.145692 |
| ACC 1h | 91.5574 | 11.0722 | 0.428366 | 0.0292593 |
| BABA 1h | 129.749 | 43.8444 | 0.552739 | 0.213515 |
| Chitin 1h | 82.4217 | 5.5396 | 0.446918 | 0.0299993 |
| Epi 1h | 121.991 | 15.1915 | 0.677585 | 0.0675479 |
| SA 1h | 104.335 | 31.8606 | 0.441852 | 0.167219 |
| Me-JA 1h | 34.4497 | 9.09025 | 0.189971 | 0.0411356 |
| Control 6h | 795.731 | 300.866 | 3.3071 | 1.21144 |
| ABA 6h | 272.945 | 37.6757 | 1.22733 | 0.116071 |
| ACC 6h | 467.803 | 107.861 | 1.86323 | 0.369675 |
| BABA 6h | 566.951 | 141.434 | 2.31751 | 0.55645 |
| Chitin 6h | 466.632 | 97.0532 | 2.05019 | 0.393354 |
| Epi 6h | 664.714 | 121.666 | 2.78351 | 0.626105 |
| SA 6h | 429.833 | 89.3092 | 2.01585 | 0.625776 |
| Me-JA 6h | 207.639 | 94.4463 | 0.74285 | 0.505806 |
Source Transcript PGSC0003DMT400061950 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G43670.1 | +2 | 1e-161 | 382 | 196/229 (86%) | Inositol monophosphatase family protein | chr1:16468184-16470347 FORWARD LENGTH=341 |
