Probe CUST_37050_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37050_PI426222305 | JHI_St_60k_v1 | DMT400061949 | ATTCTGTCTGCACTTGACTTAGTTCCAGAGAAGATACACGAACGCTCTCCTATATTTCTT |
All Microarray Probes Designed to Gene DMG400024109
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37050_PI426222305 | JHI_St_60k_v1 | DMT400061949 | ATTCTGTCTGCACTTGACTTAGTTCCAGAGAAGATACACGAACGCTCTCCTATATTTCTT |
| CUST_37042_PI426222305 | JHI_St_60k_v1 | DMT400061950 | CAAGTACGCTTATTGGAAAGCAATAGCTAATAAGTTTGTTTCCCATGTTCATGATATCCT |
| CUST_37033_PI426222305 | JHI_St_60k_v1 | DMT400061951 | GCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTTTTT |
| CUST_37071_PI426222305 | JHI_St_60k_v1 | DMT400061953 | GCGTTTTGTTCTATTATTAGAACGTGACACTCGCAGTAATGTTATTATGCAGGCACTTGA |
| CUST_37028_PI426222305 | JHI_St_60k_v1 | DMT400061952 | GTTGCACAATAGCTGCTGTGACGCGATAATACGTTCACATTACTGGCTTGTTCTAACTTT |
Microarray Signals from CUST_37050_PI426222305
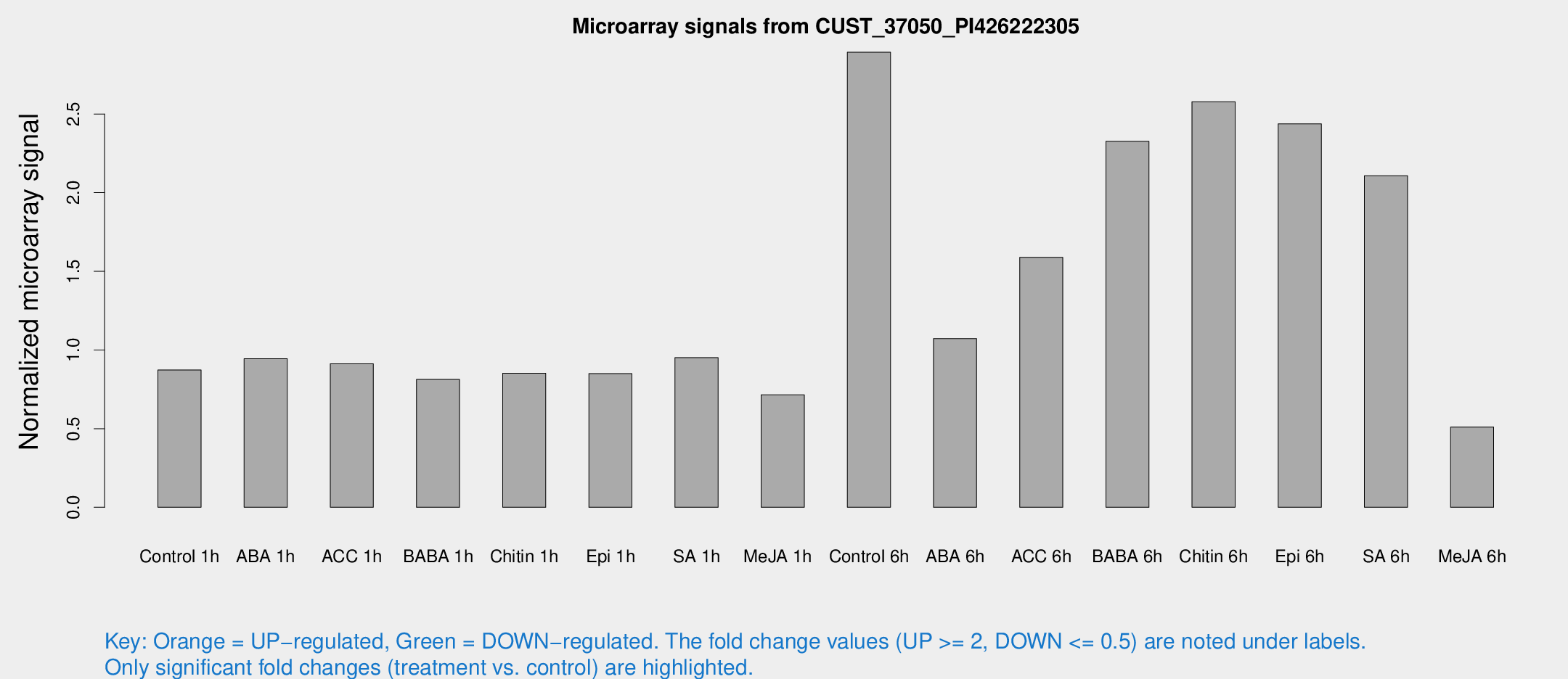
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7680.44 | 1235.42 | 0.872824 | 0.0866293 |
| ABA 1h | 7326.57 | 1164.23 | 0.943975 | 0.0811138 |
| ACC 1h | 8071.83 | 585.222 | 0.912634 | 0.0526925 |
| BABA 1h | 7402.78 | 1996.53 | 0.812853 | 0.193107 |
| Chitin 1h | 6590.16 | 407.866 | 0.852105 | 0.0557731 |
| Epi 1h | 6335.93 | 478.351 | 0.849787 | 0.0490642 |
| SA 1h | 8732.42 | 1869.6 | 0.951567 | 0.145216 |
| Me-JA 1h | 5088.52 | 699.278 | 0.714809 | 0.0502572 |
| Control 6h | 26359.4 | 6176.4 | 2.89263 | 0.518844 |
| ABA 6h | 9771.94 | 564.211 | 1.0728 | 0.066011 |
| ACC 6h | 16551.3 | 3813.19 | 1.58903 | 0.228967 |
| BABA 6h | 23403.4 | 5306.87 | 2.3266 | 0.524976 |
| Chitin 6h | 24037.3 | 3688.45 | 2.5774 | 0.303347 |
| Epi 6h | 24157.9 | 4014.94 | 2.43778 | 0.409553 |
| SA 6h | 18451.5 | 3145.17 | 2.10775 | 0.154281 |
| Me-JA 6h | 5383.67 | 2262.58 | 0.510099 | 0.230104 |
Source Transcript PGSC0003DMT400061949 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G43670.1 | +1 | 0.0 | 561 | 280/347 (81%) | Inositol monophosphatase family protein | chr1:16468184-16470347 FORWARD LENGTH=341 |
