Probe CUST_31504_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31504_PI426222305 | JHI_St_60k_v1 | DMT400042617 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
All Microarray Probes Designed to Gene DMG400016530
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31557_PI426222305 | JHI_St_60k_v1 | DMT400042621 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31423_PI426222305 | JHI_St_60k_v1 | DMT400042616 | CCACTGTATTCTTATCGCATATTTTGCTCTTTAACAGGTCATAAATTTTTTAGTCGGCCT |
| CUST_31425_PI426222305 | JHI_St_60k_v1 | DMT400042618 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31440_PI426222305 | JHI_St_60k_v1 | DMT400042620 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31504_PI426222305 | JHI_St_60k_v1 | DMT400042617 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
Microarray Signals from CUST_31504_PI426222305
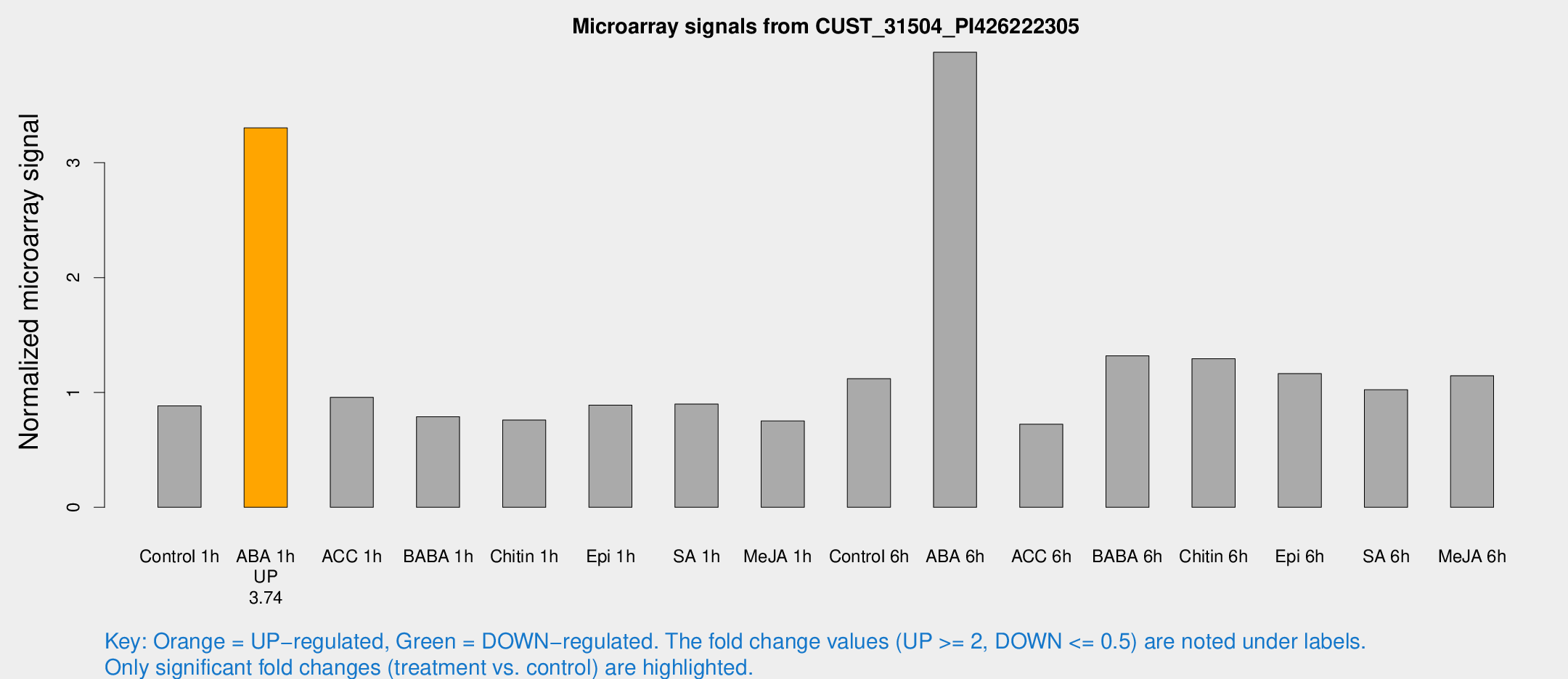
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 270.654 | 39.1013 | 0.88313 | 0.0669067 |
| ABA 1h | 891.053 | 112.613 | 3.30334 | 0.443508 |
| ACC 1h | 307.307 | 57.7142 | 0.957773 | 0.121803 |
| BABA 1h | 228.389 | 13.5912 | 0.788714 | 0.0925962 |
| Chitin 1h | 209.431 | 29.3516 | 0.759641 | 0.0894796 |
| Epi 1h | 233.451 | 22.7734 | 0.889289 | 0.0859787 |
| SA 1h | 279.347 | 23.9299 | 0.898716 | 0.114817 |
| Me-JA 1h | 184.301 | 11.115 | 0.751437 | 0.0652597 |
| Control 6h | 392.526 | 134.134 | 1.11958 | 0.388947 |
| ABA 6h | 1272.63 | 96.9371 | 3.96191 | 0.269271 |
| ACC 6h | 254.029 | 34.9078 | 0.723404 | 0.0431747 |
| BABA 6h | 443.512 | 25.8693 | 1.3182 | 0.0768409 |
| Chitin 6h | 415.471 | 28.5554 | 1.29328 | 0.0806072 |
| Epi 6h | 402.306 | 55.69 | 1.16376 | 0.23964 |
| SA 6h | 313.738 | 56.6321 | 1.02354 | 0.123274 |
| Me-JA 6h | 359.314 | 79.1299 | 1.14536 | 0.156701 |
Source Transcript PGSC0003DMT400042617 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G67300.2 | +2 | 1e-39 | 146 | 69/95 (73%) | Major facilitator superfamily protein | chr1:25193832-25196751 REVERSE LENGTH=494 |
