Probe CUST_31557_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31557_PI426222305 | JHI_St_60k_v1 | DMT400042621 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
All Microarray Probes Designed to Gene DMG400016530
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31557_PI426222305 | JHI_St_60k_v1 | DMT400042621 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31423_PI426222305 | JHI_St_60k_v1 | DMT400042616 | CCACTGTATTCTTATCGCATATTTTGCTCTTTAACAGGTCATAAATTTTTTAGTCGGCCT |
| CUST_31425_PI426222305 | JHI_St_60k_v1 | DMT400042618 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31440_PI426222305 | JHI_St_60k_v1 | DMT400042620 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31504_PI426222305 | JHI_St_60k_v1 | DMT400042617 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
Microarray Signals from CUST_31557_PI426222305
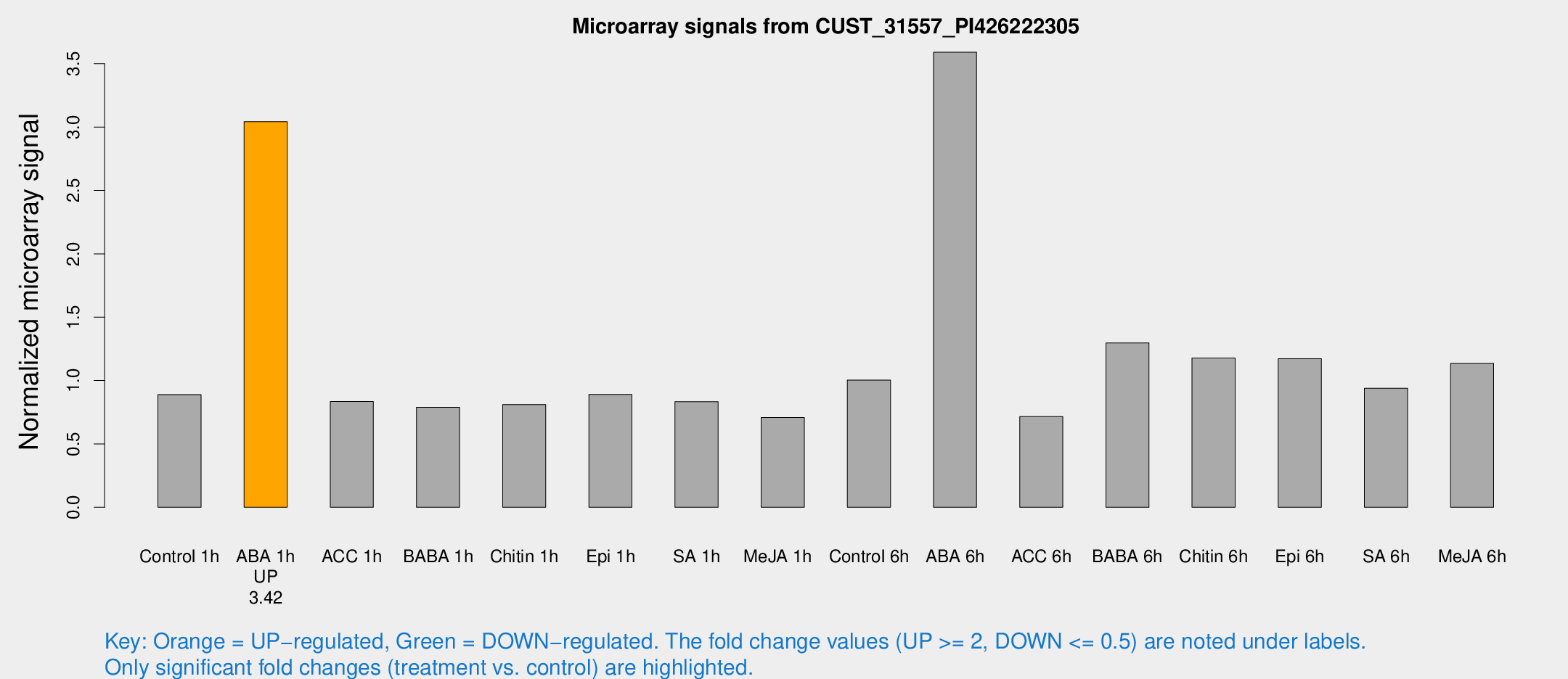
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 279.038 | 25.9514 | 0.889653 | 0.0526049 |
| ABA 1h | 840.949 | 61.6367 | 3.04228 | 0.205056 |
| ACC 1h | 278.861 | 58.6464 | 0.834268 | 0.122899 |
| BABA 1h | 237.278 | 14.4049 | 0.789271 | 0.133236 |
| Chitin 1h | 233.818 | 41.8305 | 0.810669 | 0.132907 |
| Epi 1h | 242.124 | 24.0012 | 0.890842 | 0.0855121 |
| SA 1h | 274.147 | 47.9603 | 0.8329 | 0.179966 |
| Me-JA 1h | 183.632 | 25.1123 | 0.70904 | 0.1617 |
| Control 6h | 369.241 | 126.406 | 1.00409 | 0.38087 |
| ABA 6h | 1190.18 | 69.042 | 3.59062 | 0.250523 |
| ACC 6h | 260.967 | 39.7708 | 0.716392 | 0.0431363 |
| BABA 6h | 454.183 | 36.0534 | 1.29756 | 0.120505 |
| Chitin 6h | 392.787 | 35.7362 | 1.17801 | 0.10902 |
| Epi 6h | 418.671 | 56.5443 | 1.17269 | 0.22283 |
| SA 6h | 290.371 | 21.9606 | 0.939383 | 0.0852348 |
| Me-JA 6h | 374.964 | 91.5116 | 1.13519 | 0.203764 |
Source Transcript PGSC0003DMT400042621 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79820.1 | +1 | 0.0 | 626 | 326/477 (68%) | Major facilitator superfamily protein | chr1:30022581-30026771 REVERSE LENGTH=495 |
