Probe CUST_31440_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31440_PI426222305 | JHI_St_60k_v1 | DMT400042620 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
All Microarray Probes Designed to Gene DMG400016530
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31557_PI426222305 | JHI_St_60k_v1 | DMT400042621 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31423_PI426222305 | JHI_St_60k_v1 | DMT400042616 | CCACTGTATTCTTATCGCATATTTTGCTCTTTAACAGGTCATAAATTTTTTAGTCGGCCT |
| CUST_31425_PI426222305 | JHI_St_60k_v1 | DMT400042618 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31440_PI426222305 | JHI_St_60k_v1 | DMT400042620 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31504_PI426222305 | JHI_St_60k_v1 | DMT400042617 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
Microarray Signals from CUST_31440_PI426222305
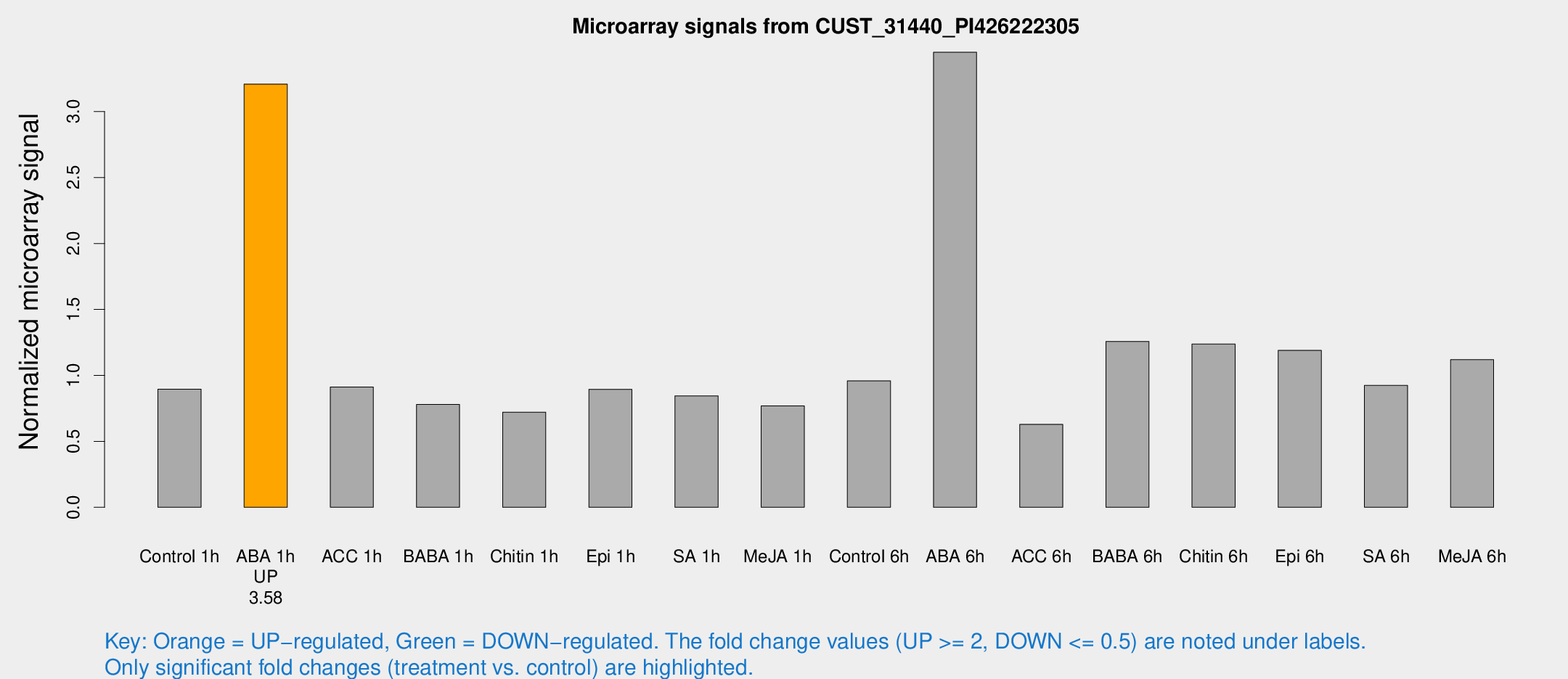
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 296.059 | 39.6554 | 0.895632 | 0.0639497 |
| ABA 1h | 929.1 | 86.8922 | 3.20842 | 0.283264 |
| ACC 1h | 309.011 | 40.1102 | 0.911323 | 0.056953 |
| BABA 1h | 244.71 | 14.5753 | 0.779789 | 0.117241 |
| Chitin 1h | 223.385 | 57.2234 | 0.721414 | 0.188741 |
| Epi 1h | 255.477 | 31.4238 | 0.894486 | 0.106865 |
| SA 1h | 285.428 | 32.7966 | 0.844273 | 0.146045 |
| Me-JA 1h | 204.837 | 16.1992 | 0.768381 | 0.112219 |
| Control 6h | 371.097 | 131.69 | 0.958018 | 0.376433 |
| ABA 6h | 1200.8 | 111.953 | 3.44988 | 0.293148 |
| ACC 6h | 244.991 | 52.9383 | 0.628846 | 0.0538342 |
| BABA 6h | 459.208 | 33.1701 | 1.25756 | 0.119658 |
| Chitin 6h | 429.35 | 29.5114 | 1.23732 | 0.0957428 |
| Epi 6h | 451.248 | 78.8477 | 1.18983 | 0.326799 |
| SA 6h | 298.274 | 22.2652 | 0.924286 | 0.111084 |
| Me-JA 6h | 384.185 | 87.5438 | 1.11887 | 0.196054 |
Source Transcript PGSC0003DMT400042620 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79820.1 | +1 | 8e-174 | 380 | 191/283 (67%) | Major facilitator superfamily protein | chr1:30022581-30026771 REVERSE LENGTH=495 |
