Probe CUST_31425_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31425_PI426222305 | JHI_St_60k_v1 | DMT400042618 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
All Microarray Probes Designed to Gene DMG400016530
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31557_PI426222305 | JHI_St_60k_v1 | DMT400042621 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31423_PI426222305 | JHI_St_60k_v1 | DMT400042616 | CCACTGTATTCTTATCGCATATTTTGCTCTTTAACAGGTCATAAATTTTTTAGTCGGCCT |
| CUST_31425_PI426222305 | JHI_St_60k_v1 | DMT400042618 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31440_PI426222305 | JHI_St_60k_v1 | DMT400042620 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
| CUST_31504_PI426222305 | JHI_St_60k_v1 | DMT400042617 | GATACTGGTAGCTGCTTTGCTGTAACAAGAAATATTGGTAGTATTGTAGTACACAATTGA |
Microarray Signals from CUST_31425_PI426222305
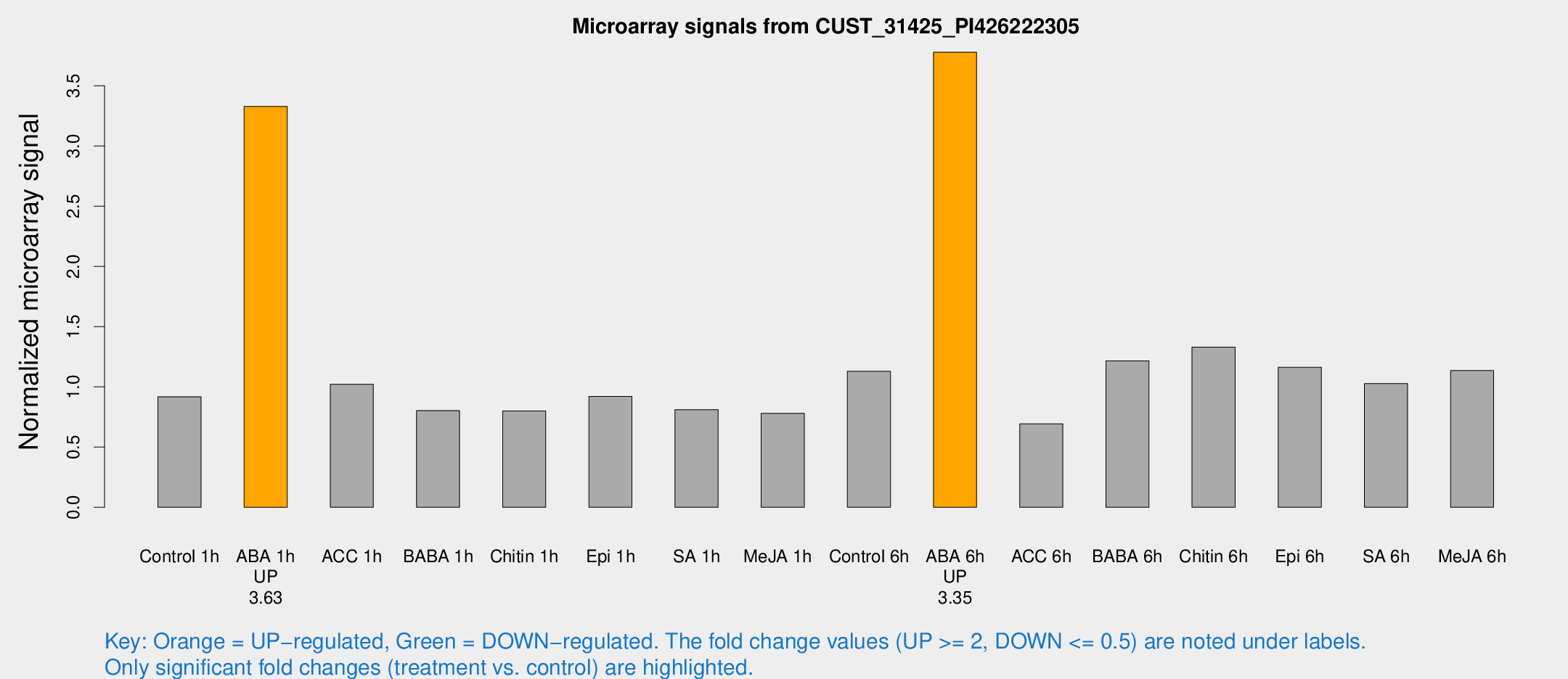
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 281.268 | 33.2037 | 0.917894 | 0.0539939 |
| ABA 1h | 903.213 | 111.674 | 3.3287 | 0.366087 |
| ACC 1h | 327.085 | 56.3463 | 1.02201 | 0.108704 |
| BABA 1h | 233.826 | 13.9015 | 0.802539 | 0.0863372 |
| Chitin 1h | 226.824 | 47.3223 | 0.800299 | 0.154773 |
| Epi 1h | 243.608 | 26.0228 | 0.920521 | 0.0896755 |
| SA 1h | 255.802 | 31.7167 | 0.811395 | 0.143817 |
| Me-JA 1h | 193.452 | 14.4453 | 0.780144 | 0.118882 |
| Control 6h | 379.136 | 107.387 | 1.12941 | 0.289867 |
| ABA 6h | 1226.95 | 126.499 | 3.77907 | 0.385821 |
| ACC 6h | 249.868 | 51.2945 | 0.693576 | 0.0413779 |
| BABA 6h | 411.483 | 24.0366 | 1.21595 | 0.0709638 |
| Chitin 6h | 429.724 | 30.1252 | 1.3303 | 0.117011 |
| Epi 6h | 410.55 | 72.0263 | 1.16279 | 0.321381 |
| SA 6h | 308.988 | 24.2696 | 1.028 | 0.0696403 |
| Me-JA 6h | 375.615 | 110.48 | 1.13566 | 0.272428 |
Source Transcript PGSC0003DMT400042618 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79820.1 | +3 | 1e-174 | 422 | 223/328 (68%) | Major facilitator superfamily protein | chr1:30022581-30026771 REVERSE LENGTH=495 |
