Probe CUST_25480_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25480_PI426222305 | JHI_St_60k_v1 | DMT400034701 | TTTATCTTGATGAAAAGAACCGCCCTCTGTTAATTCACTGCAAAAGAGGAAAGATGAACG |
All Microarray Probes Designed to Gene DMG402013333
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25480_PI426222305 | JHI_St_60k_v1 | DMT400034701 | TTTATCTTGATGAAAAGAACCGCCCTCTGTTAATTCACTGCAAAAGAGGAAAGATGAACG |
| CUST_25459_PI426222305 | JHI_St_60k_v1 | DMT400034700 | TGGTTGTTGCTGTTAAAATCCCAACCTTTCTACTCGAAGAGTTGTGAGTTTGTTAAAAAA |
| CUST_25316_PI426222305 | JHI_St_60k_v1 | DMT400034702 | AAGAGGAAAGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAG |
| CUST_25464_PI426222305 | JHI_St_60k_v1 | DMT400034703 | TTTATCTTGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAGG |
Microarray Signals from CUST_25480_PI426222305
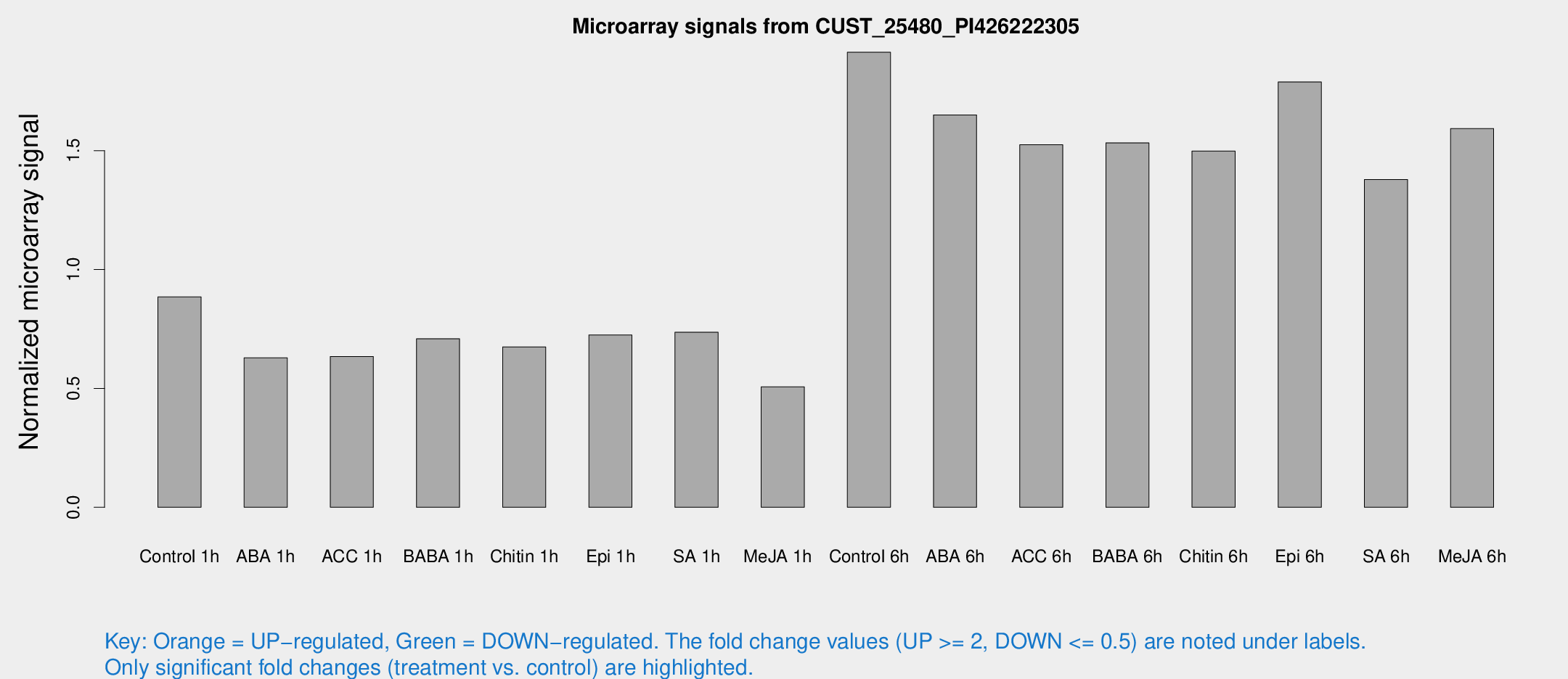
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 385.261 | 43.3753 | 0.884856 | 0.0518296 |
| ABA 1h | 242.713 | 29.7847 | 0.628749 | 0.0958623 |
| ACC 1h | 299.462 | 69.168 | 0.634514 | 0.126602 |
| BABA 1h | 311.171 | 67.3223 | 0.709042 | 0.103742 |
| Chitin 1h | 262.609 | 20.9215 | 0.674709 | 0.0400745 |
| Epi 1h | 270.683 | 16.0864 | 0.724827 | 0.0429786 |
| SA 1h | 332.606 | 49.9341 | 0.736489 | 0.0636544 |
| Me-JA 1h | 180.207 | 20.9539 | 0.506513 | 0.0383785 |
| Control 6h | 918.418 | 290.902 | 1.91415 | 0.528971 |
| ABA 6h | 791.007 | 156.668 | 1.65058 | 0.316619 |
| ACC 6h | 773.877 | 133.692 | 1.52492 | 0.0884762 |
| BABA 6h | 753 | 109.831 | 1.53278 | 0.202744 |
| Chitin 6h | 694.139 | 75.5459 | 1.49855 | 0.112424 |
| Epi 6h | 886.466 | 130.253 | 1.78934 | 0.22846 |
| SA 6h | 630.661 | 156.322 | 1.37845 | 0.234359 |
| Me-JA 6h | 697.202 | 97.9671 | 1.59288 | 0.104544 |
Source Transcript PGSC0003DMT400034701 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05000.2 | +2 | 1e-38 | 142 | 65/86 (76%) | Phosphotyrosine protein phosphatases superfamily protein | chr1:1425660-1428393 FORWARD LENGTH=247 |
