Probe CUST_25459_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25459_PI426222305 | JHI_St_60k_v1 | DMT400034700 | TGGTTGTTGCTGTTAAAATCCCAACCTTTCTACTCGAAGAGTTGTGAGTTTGTTAAAAAA |
All Microarray Probes Designed to Gene DMG402013333
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25480_PI426222305 | JHI_St_60k_v1 | DMT400034701 | TTTATCTTGATGAAAAGAACCGCCCTCTGTTAATTCACTGCAAAAGAGGAAAGATGAACG |
| CUST_25459_PI426222305 | JHI_St_60k_v1 | DMT400034700 | TGGTTGTTGCTGTTAAAATCCCAACCTTTCTACTCGAAGAGTTGTGAGTTTGTTAAAAAA |
| CUST_25316_PI426222305 | JHI_St_60k_v1 | DMT400034702 | AAGAGGAAAGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAG |
| CUST_25464_PI426222305 | JHI_St_60k_v1 | DMT400034703 | TTTATCTTGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAGG |
Microarray Signals from CUST_25459_PI426222305
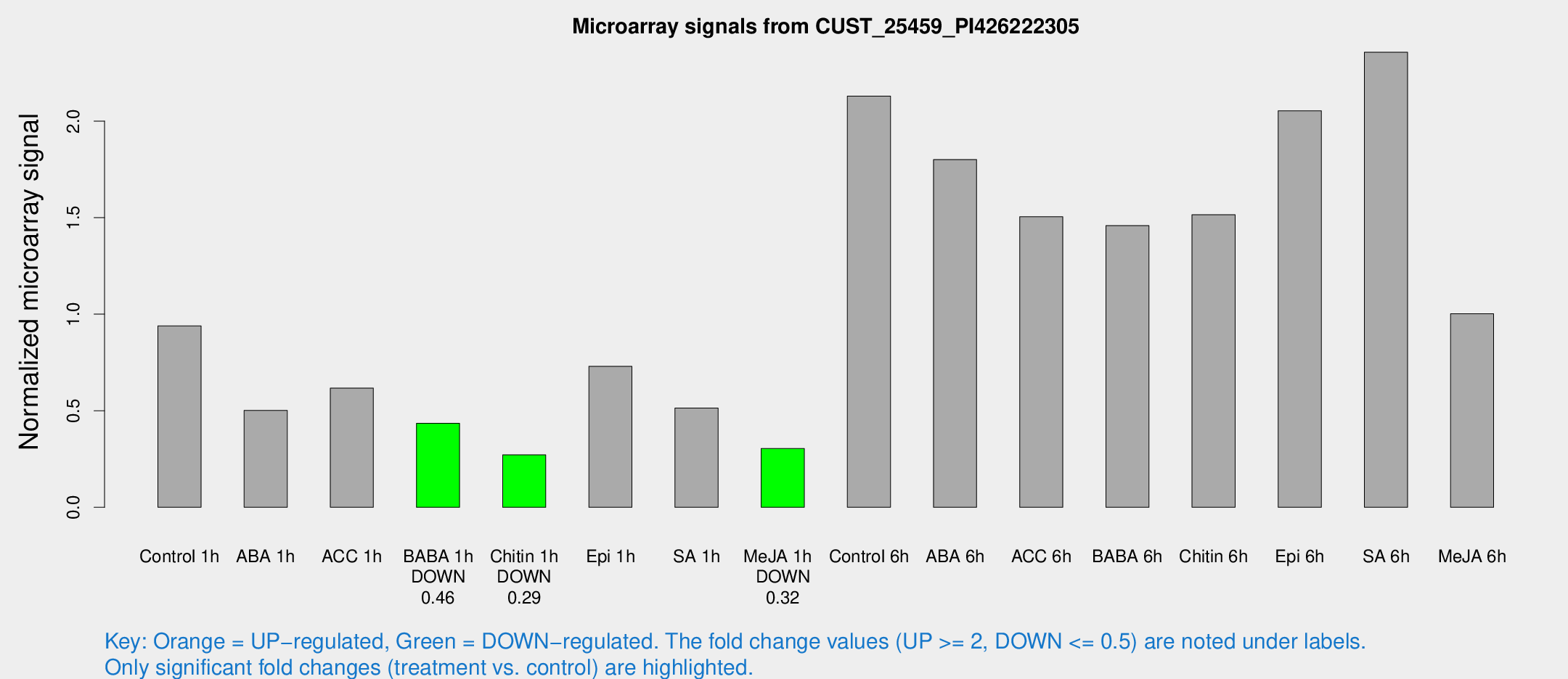
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.5154 | 3.56714 | 0.939331 | 0.150935 |
| ABA 1h | 12.0746 | 4.6564 | 0.50178 | 0.178959 |
| ACC 1h | 17.9287 | 6.62232 | 0.616938 | 0.257173 |
| BABA 1h | 10.6935 | 3.57938 | 0.434683 | 0.170433 |
| Chitin 1h | 5.8385 | 3.41043 | 0.271277 | 0.157285 |
| Epi 1h | 15.7296 | 3.4116 | 0.729862 | 0.182728 |
| SA 1h | 14.8916 | 5.36245 | 0.513531 | 0.234773 |
| Me-JA 1h | 5.931 | 3.45517 | 0.304099 | 0.176208 |
| Control 6h | 58.2267 | 20.7229 | 2.12953 | 0.705597 |
| ABA 6h | 67.5995 | 33.7074 | 1.80061 | 1.69425 |
| ACC 6h | 41.5006 | 4.88649 | 1.50473 | 0.312308 |
| BABA 6h | 43.585 | 14.6023 | 1.45841 | 0.575367 |
| Chitin 6h | 39.427 | 6.83596 | 1.51501 | 0.206994 |
| Epi 6h | 61.7037 | 21.9216 | 2.0528 | 0.705418 |
| SA 6h | 56.6753 | 8.8571 | 2.35647 | 0.269882 |
| Me-JA 6h | 25.9432 | 7.05317 | 1.00196 | 0.239623 |
Source Transcript PGSC0003DMT400034700 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05000.2 | +1 | 5e-26 | 107 | 58/115 (50%) | Phosphotyrosine protein phosphatases superfamily protein | chr1:1425660-1428393 FORWARD LENGTH=247 |
